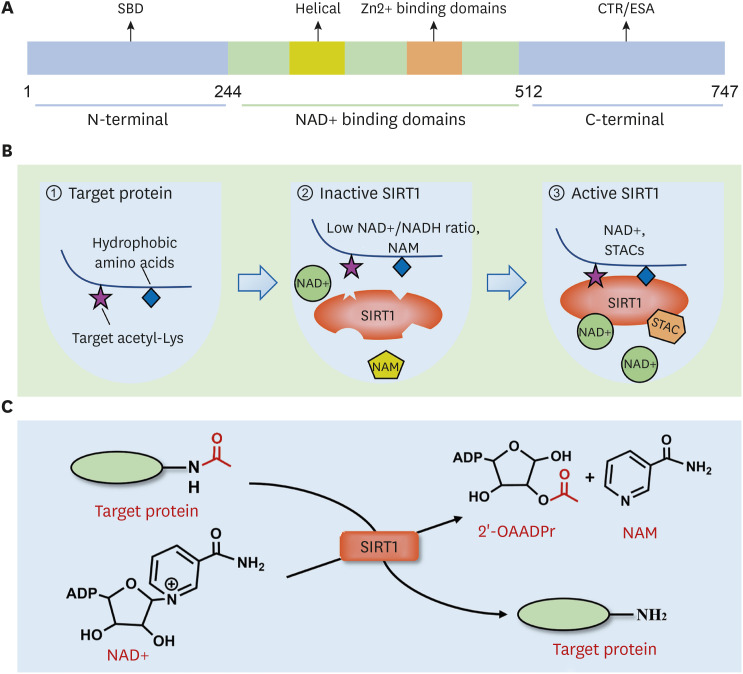
Figure 1. Molecular structure and biological functional characteristics of SIRT1. (A) SIRT1 has an evolutionarily conserved NAD+-dependent catalytic core domain (244–512 residues), unique N-terminal (513–747 residues), and C-terminal (1–180 residues) sequences. SBD locates on N-terminal, and CTR/ESA locates on C-terminal. (B) SIRT1 prefers specific hydrophobic amino acids near the target Lys residue for substrate recognition (panel 1). Low NAD+/NADH ratio and NAM weaken the activity of SIRT1 (panel 2). STACs activate SIRT1 by combing with the SBD, increasing the catalytic activity of SIRT1 (panel 3). (C) The deacetylation mechanism is mediated by SIRT1. The acetyl group of the substrate is transferred to the ADP ribosyl part of NAD+, while an NAD+ molecule is split into 1 NAM and 1 2-OAADPr.
SBD, sirtuins-activating compounds binding domain; STAC, sirtuins-activating compound; CTR, C-terminal regulatory segment.