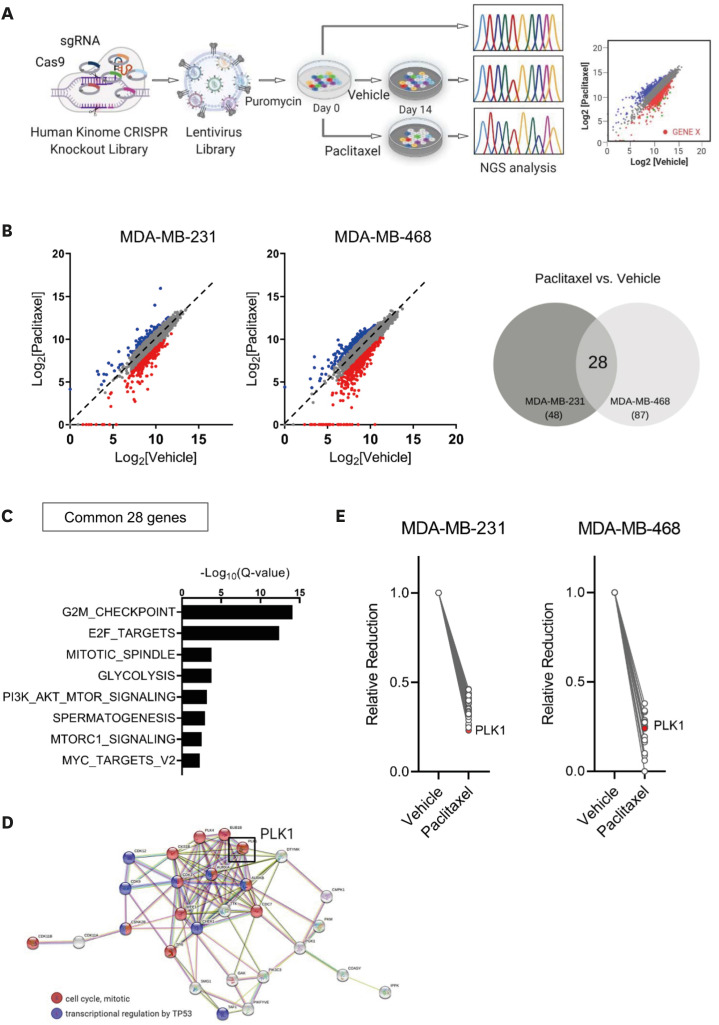
Figure 1. Human kinome CRISPR/Cas9 screening for the identification of candidate therapeutic target genes related to paclitaxel resistance in breast cancer.
(A) Schematic illustration showing the human kinome CRISPR/Cas9 knockout screens used to identify genes associated with PTX resistance in MDA-MB-231 and MDA-MB-468 cells. (B) Scatterplots of normalized sgRNA counts for PTX- versus vehicle-treatment at day 14 (left panel). Red dots show that the sgRNA frequencies were depleted in PTX-treated cells (log2 [fold change] ≤ −1). Venn diagram of 28 genes that overlapped in MDA-MB-231 and MDA-MB-468 cells (right panel). (C) Significantly enriched gene sets (FDR Q < 0.01) for the 28 genes from the PTX-treated group. (D) STRING network analysis of the 28 genes depleted in the PTX-treated group. Genes involved in cell cycle progression and transcriptional regulation by TP53 are indicated in red and blue, respectively. PLK1 indicated by a black box is a one of the hub nodes in the network. (E) Changes in the normalized sgRNA counts in key hub genes between the vehicle- and PTX-treated groups.
FDR = false discovery rate; NGS = next-generation sequencing; PLK1 = polo-like kinase 1; PTX = paclitaxel; sgRNA = single-guide RNA.