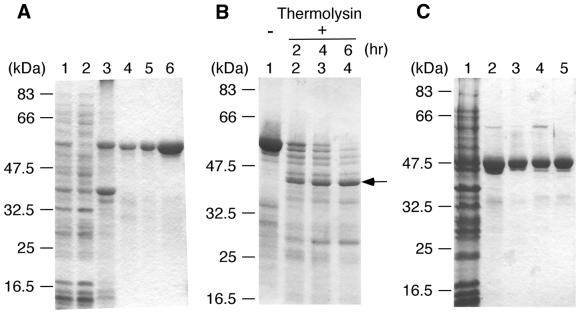
Figure 2.
Purification of ttRecJ and cd-ttRecJ and limited proteolysis of ttRecJ. (A) Purification of ttRecJ. Lane 1, total cell extract; lane 2, supernatant of the total cell extract; lane 3, extract from the pellet; lane 4, Ni2+–nitriloacetate–agarose; lane 5, refolding fraction; lane 6, MonoQ HR5/5. (B) Limited proteolysis of ttRecJ with thermolysin. An aliquot of 10 µM ttRecJ was reacted with 0.5 µg/ml thermolysin in 20 mM Tris–HCl buffer, pH 8.0, containing 2 mM CaCl2. This reaction mixture was incubated at 25°C for the indicated incubation periods. Lane 1, ttRecJ; lanes 2–4, digests incubated for the periods indicated above each lane. The arrow indicates the core domain of ttRecJ. (C) Purification of cd-ttRecJ. Lane 1, supernatant of the total cell extract; lane 2, Ni2+–nitriloacetate–agarose; lane 3, Bio-Gel HTP gel; lane 4, MonoQ HR5/5; lane 5, HiPrep 16/60 Sephacryl S-200 HR. Each step was analyzed by staining the SDS–PAGE gel (12% w/v acrylamide) with Coomassie brilliant blue. The molecular mass markers were: MBP–paramyosin, 83 kDa; glutamic acid dehydrogenase, 62 kDa; aldolase, 47.5 kDa; triosephosphate isomerase, 32.5 kDa; β-lactoglobulin A, 25 kDa; lysozyme, 16.5 kDa.