Figure 1.
Molecular mechanisms of PQ resistance elucidated in Arabidopsis thaliana
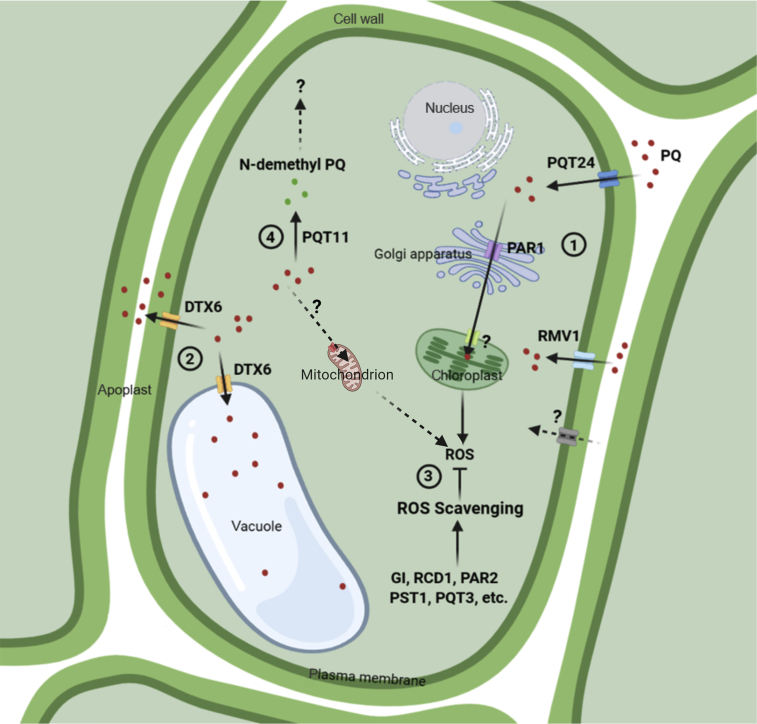
PQ is transported across the plasma membrane into the cytosol from the external environment by ABC transporters such as PDR11/PQT24 and L-amino acid transporters such as RMV1/LAT1, as well as other unidentified transporters that recognize PQ as their mimic substrate. Once inside the cell, PQ faces several fates. First, PQ is transported by PAR1/LAT4 and other unidentified transporters to its site of action, the chloroplast, where it competes for electrons from PSI and generates large amounts of ROS that are scavenged by antioxidant enzymes. Any mutations that enhance ROS-scavenging ability, such as pqt3 and pst1, would help plants to tolerate PQ. Second, PQ is exported to the vacuole and apoplast by efflux transporters such as DTX6/PQT15/RTP1. Third, PQ is catabolized to nontoxic products by plant enzymes such as PQT11/CYP86A4. Therefore, PQ resistance mechanisms revealed by Arabidopsis mutants to date include (1) impaired PQ transport (rvm1, pqt24, and par1); (2) enhanced PQ export to the vacuole and apoplast (dtx6D); (3) enhanced ROS-scavenging capability (e.g., pst1 and pqt3); and (4) enhanced metabolic detoxification of PQ (pqt11D). Unconfirmed PQ transporters and transport routes are indicated by question marks and dashed lines, respectively. Red dots represent PQ, and green dots represent N-demethyl PQ.