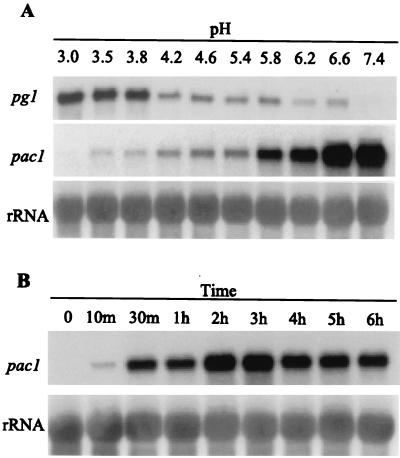
FIG. 2.
pg1 and pac1 transcript accumulation in response to different ambient pH conditions. (A) Lanes 1 to 10 show a Northern analysis of total RNA isolated from S. sclerotiorum mycelia 6 h after transfer to fresh YPSu medium buffered with citrate-phosphate buffer at the indicated pH values. Blots were probed with pac1 and then sequentially stripped and reprobed with pg1 and ribosomal DNA (rDNA) probes as indicated. (B) Northern analysis of total RNA isolated from S. sclerotiorum mycelia at various time points after transfer to fresh YPSu medium buffered with citrate-phosphate buffer at pH 7.9. Lane 1, total RNA from mycelia of the primary culture (final pH, 3.0); lanes 2 to 9, total RNA from cultures at 10 min to 6 h after transfer to pH 7.9. Blots were probed with pac1, stripped, and reprobed with the rDNA probe. To facilitate comparisons of transcript levels between various time points, the level of pac1 signal detected at the 10-min time point is set at 1X.