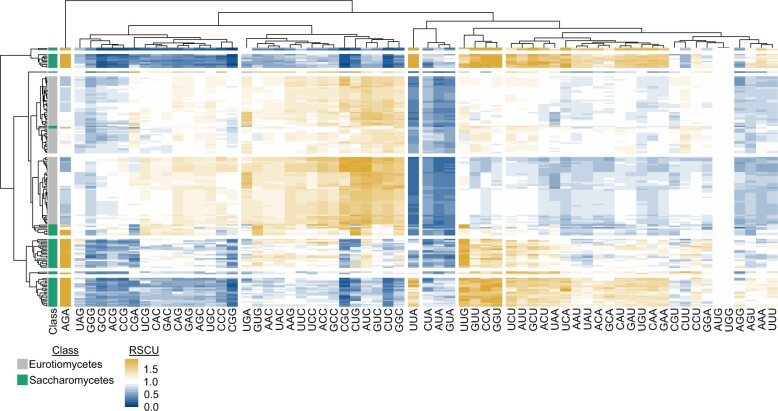
Fig. 1.
Relative synonymous codon usage across 171 fungal genomes. Relative synonymous codon usage (RSCU) was calculated from the coding sequences of 103 Eurotiomycetes (filamentous fungi) and 68 Saccharomycetes (budding yeasts) genomes obtained from NCBI. Hierarchical clustering was conducted across the fungal species (rows) and codons (columns). Eight groups of clustered rows were identified; 7 groups of clustered columns were identified. Broad differences were observed in the RSCU values of Eurotiomycetes and Saccharomycetes genomes. For example, Saccharomycetes tended to have higher RSCU values for the AGA codon, whereas Eurotiomycetes tended to have higher RSCU values for the CUG codon. To account for the use of an alternative genetic code in budding yeast genomes from the CUG-Ser1 and CUG-Ser2 lineages (Krassowski et al. 2018), the alternative yeast nuclear code—which is one of 26 alternative genetic codes incorporated into BioKIT—was used during RSCU determination. User’s may also provide their own genetic code if it is unavailable in BioKIT. This figure was made using pheatmap (Kolde 2012).