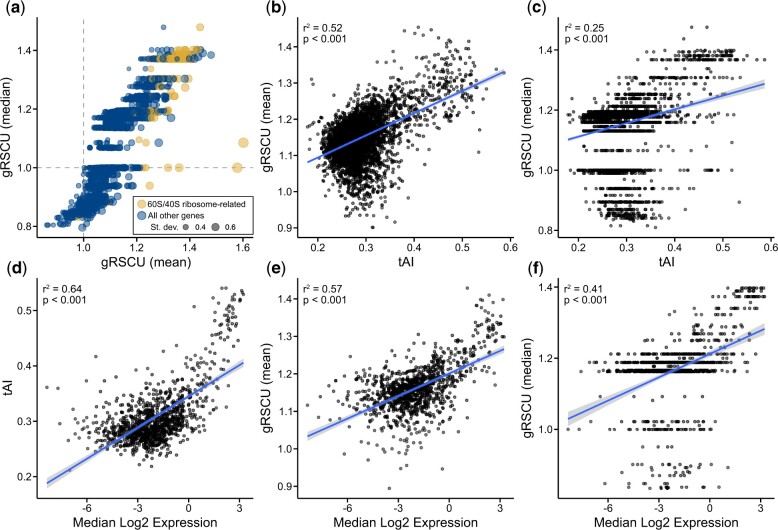
Fig. 2.
Mean gene-wise relative synonymous codon usage accurately estimates codon optimization. a) Gene-wise relative synonymous codon usage (gRSCU), the mean (x-axis) or median (y-axis) relative synonymous codon usage value per gene (based on RSCU values calculated from the entire set of protein coding genes), was calculated from the coding sequences of the model budding yeast Saccharomyces cerevisiae. b, c) In S. cerevisiae, a significant correlation was observed between tRNA adaptation index (tAI), a well-known measure of codon optimization (Sabi and Tuller 2014), and mean as well as median gRSCU (r2 = 0.52, P < 0.001 and r2 = 0.25, P < 0.001, respectively; Pearson's Correlation Coefficient). d) Using previously published data, a correlation is observed between median log2 gene expression and tAI in Saccharomyces mikatae (LaBella et al. 2019), which is evidence of tAI values being indicative of codon optimization. Comparison of mean and median gRSCU (e and f, respectively) and median log2 gene expression revealed similarly strong correlations (r2 = 0.57, P < 0.001 and r2 = 0.41, P < 0.001, respectively; Pearson’s correlation coefficient). Of note, mean gRSCU had a strong correlation to gene expression than median gRSCU. Each gene is represented by a dot. In (a), the size of each dot represents the standard deviation of RSCU values observed in the gene.