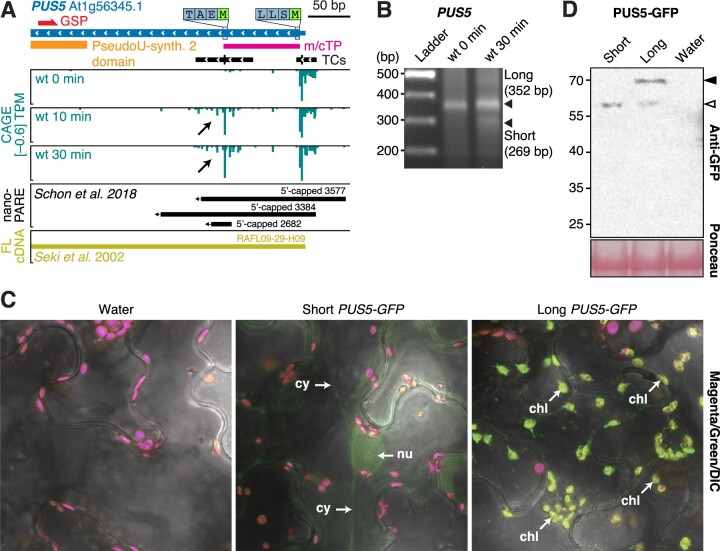
Figure 5.
Alternative TSS usage at the PUS5 locus determines the subcellular localization of the PUS5 protein. A, Genome browser view of PUS5, organized as in Figure 3A. The predicted mitochondrial/chloroplast target peptide (m/cTP) is indicated by a pink block. B, Separation of PCR fragments obtained by 5′-RACE with primers specific for PUS5, organized as in Figure 3C. The migration of the entire flg22 30 min lane in the gel with PUS5 RACE PCR products is shifted upward, explaining the apparent misalignment of the short product (269 bp) with the ladder. C, Confocal microscopy images of transiently expressed short and long PUS5 isoforms fused to GFP in N. benthamiana leaves. Signals in magenta and green channels are overlaid with the differential interference contrast microscopy images. Nu, nucleus; cy, cytoplasm; chl, chloroplast. Images from the individual channels are shown in Supplemental Figure S4 to support the identification of structures harboring GFP signal in long PUS5-GFP as chloroplasts, and to support the conclusion that signal in the green channel from chloroplasts obtained with cells expressing short PUS5-GFP does not exceed that obtained with cells that do not express GFP. D, Immunoblot of PUS5-GFP isoforms transiently expressed in N. benthamiana leaves. Protein extracts were prepared at 2 days postinfiltration and analyzed by immunoblotting using anti-GFP antibody. White arrow indicates the short/processed isoform. Black arrow indicates the long isoform. Ponceau staining of the membrane is shown as loading control.