Figure 1.
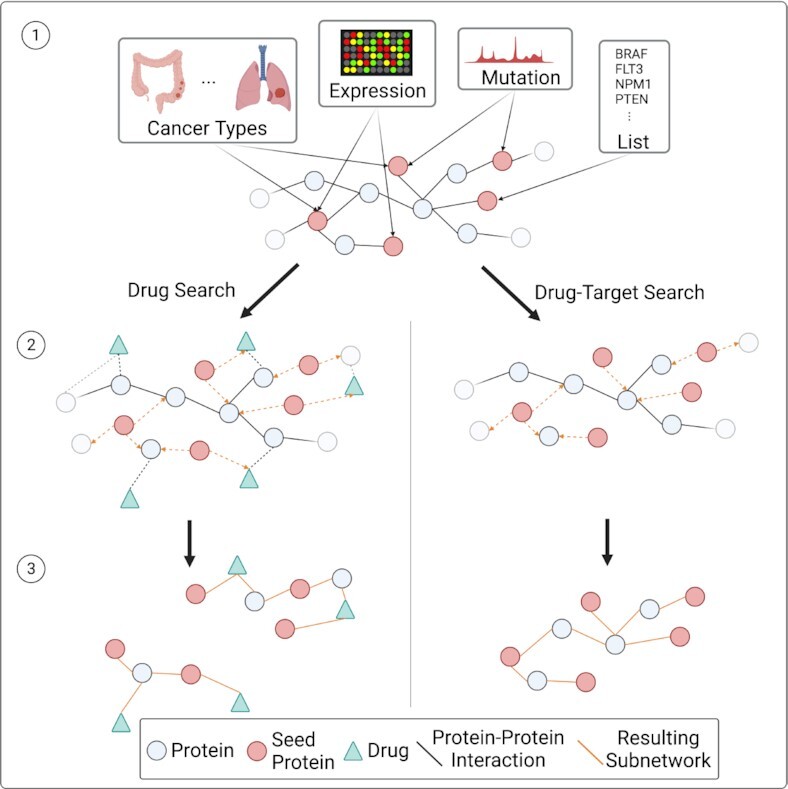
Seed genes for drug target and drug search can be selected from integrated cancer driver databases, via mutation or expression data from TCGA and GTEx using user-defined thresholds, or by uploading a list of genes (1). Based on the seed genes, putative drugs or drug target candidates are identified using a broad selection of network medicine algorithms that traverse the human gene-drug interactome (2). While the drug search reports drugs in proximity to the seed genes, the drug target search returns genes interacting with the disease module spanned by the seed genes (3).
