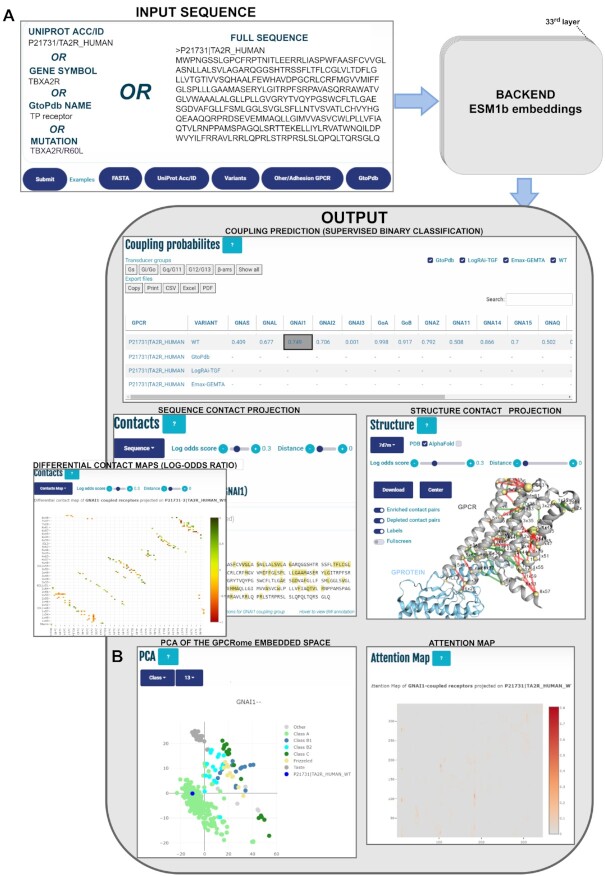
Figure 1.
(A) Workflow of PRECOGx web application. The input can be provided in different formats, including: protein identifiers (UniProt identifiers, accessions, gene symbols or GtoPdb official nomenclatures), mutations in the format protein identifier/aa substitution (e.g. MC1R/D294H) or FASTA sequences. In either cases, the corresponding sequences are used to create ESM embeddings which are in turn employed in different backend processes to generate PRECOGx output. The latter is presented in a multi-panel view including: summary table of the predicted and known couplings, predicted differential contacts mapped in 1D (Sequence panel), 2D (Contact panel) or 3D (Structure panel), attention maps and (B) K-means clustering based on GPCR class of the entire GPCRome projected along the first two components of the PCA. The number of clusters was set to 6 (possible variables: Class A, Class B1, Class B2, Class C, Frizzled, and Taste).