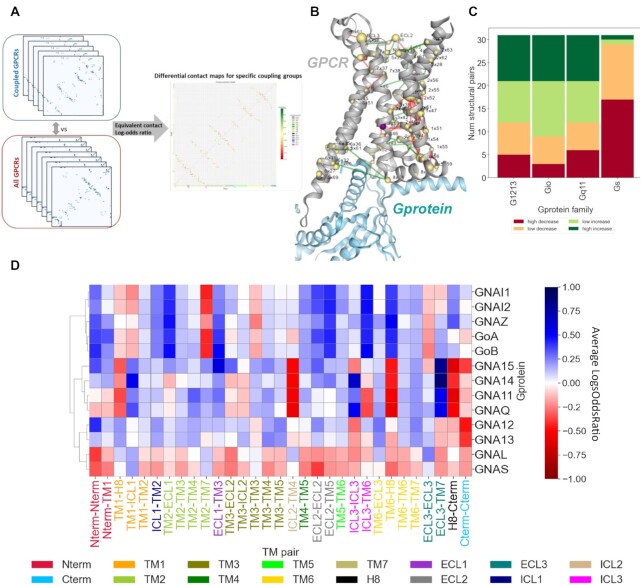
Figure 3.
Differential contact analysis: (A) Calculation of predicted contacts of each GPCR using the function predict_contacts in the ESM library and grouping GPCR based on G protein binding specificity to compute differential contact maps (log-odds ratio); (B) Projection of transducer family-specific predicted contacts on a structure in PRECOGx application (PDB 7JOZ); (C) Number of aggregated enriched(increase) or depleted(decreased) secondary structure element pairs for each Gprotein transducer family; (D) Contact enrichment signature of each Gprotein transducer family on the basis of secondary structure element.