Figure 1.
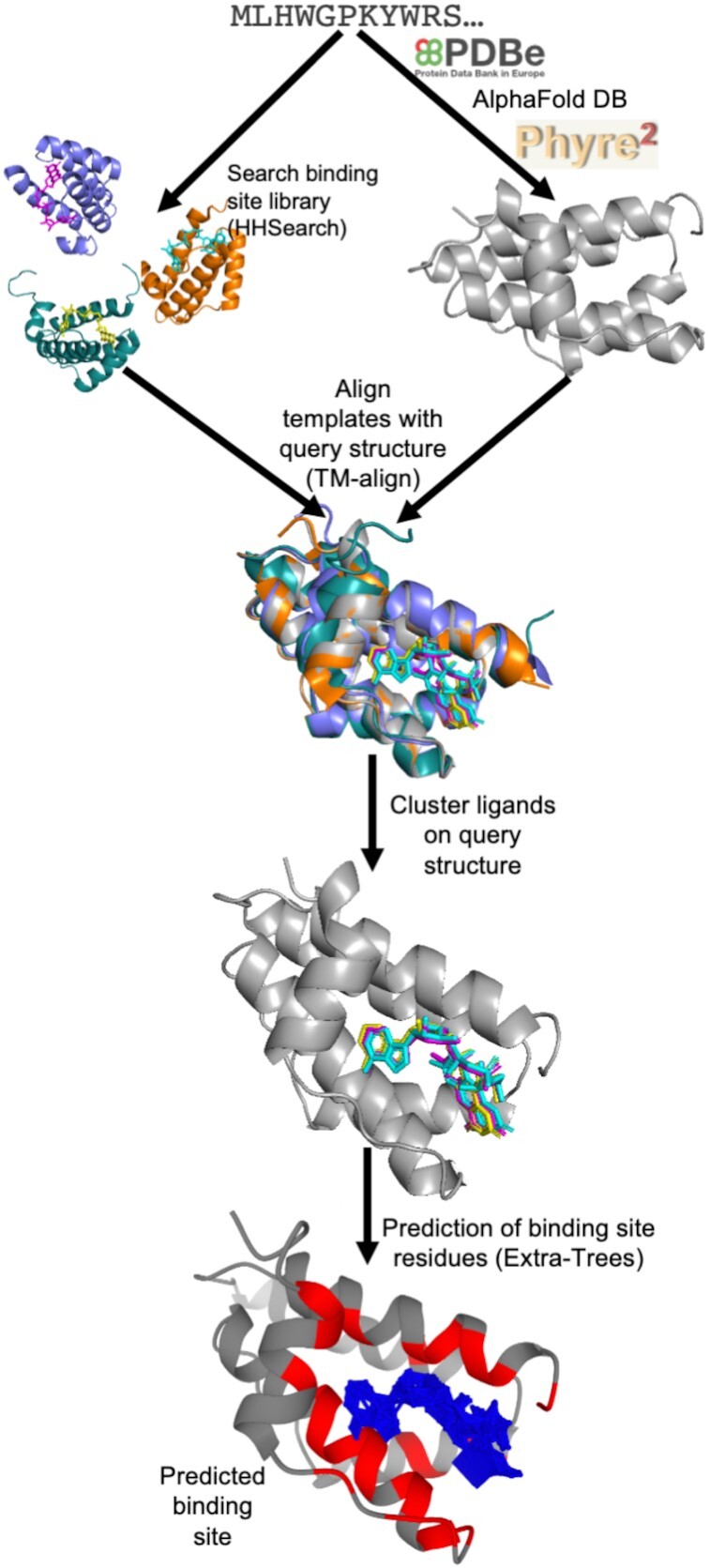
An overview of the 3DLigandSite method. Users submit either a protein sequence or structure. Where sequences are submitted, the PDBe and AlphaFold DB are searched for a matching structure; where one is not available, Phyre2 is used to model the 3D structure. HHSearch is used to search a sequence library of protein structures with ligands bound. Hits from this search are aligned with the structure of query protein, and the ligands from these structures are clustered. Each cluster of ligands represents a potential binding site in the query protein. A machine learning classifier is used to predict which of the residues around the cluster are likely to form part of a binding site.
