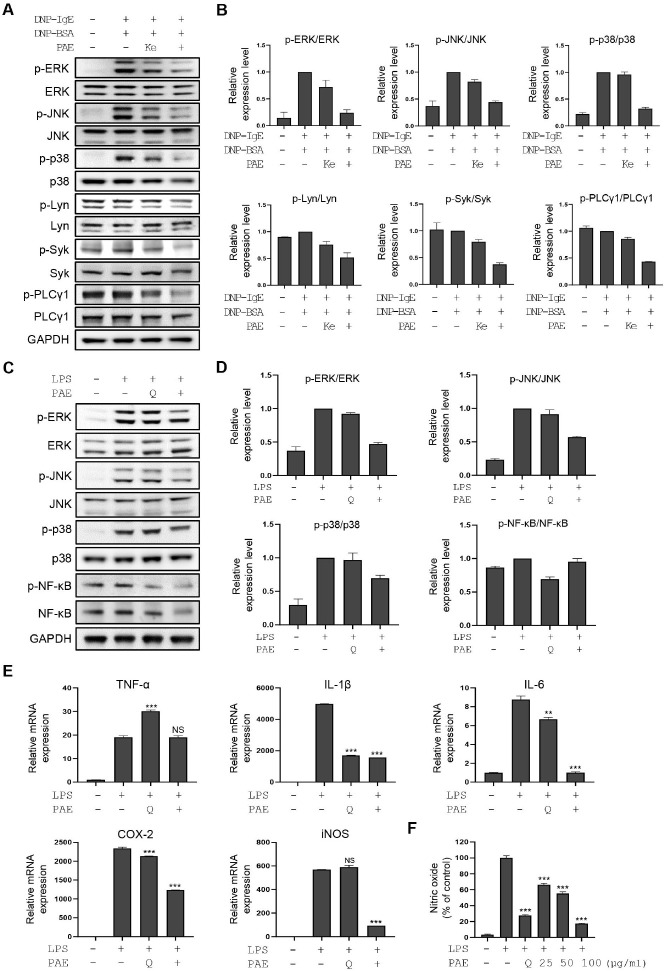
Fig. 2.
PAE shows inhibitory activity in RBL-2H3 cells via perturbation of the FcεRI pathway and in RAW264.7 cells via perturbation of the TLR4 signaling pathway. (A) Western blot analysis of the FcεRI signaling molecules, including ERK, phospho-ERK, JNK, phospho-JNK, p38, phospho-p38, Lyn, phospho-Lyn, Syk, phospho-Syk, PLC γ1, and phospho-PLC γ1, is performed in DNP-BSA/IgE-stimulated RBL-2H3 cells in the presence and absence of PAE. Ketotifen is used as a positive control (100 μM). (B) Quantification of the relative band intensities using ImageJ software. Data are expressed as means ± SD (n = 2). (C) Western blot analysis of TLR4 signaling molecules including ERK, phospho-ERK, JNK, phospho-JNK, p38, phospho-p38, NF-kB, and phos-pho-NF-kB is performed in LPS-stimulated RAW264.7 cells after PAE treatment. Quercetin is used as a positive control (100 μM). (D) Quan-tification of the relative band intensities. Data are expressed as means ± SD (n = 2). (E) The qRT-PCR analysis of TNF-α, IL-1β, IL-6, COX-2, and iNOS was performed in LPS-stimulated RAW264.7 cells after PAE treatment. (F) Nitric oxide production is measured using Griess reagent in LPS-stimulated RAW264.7 cells after PAE treatment in a dose-dependent manner. RBL-2H3 cells were co-treated with PAE and DNP-BSA for 2 hours. RAW264.7 cells were co-treated with PAE (100 μg/ml) and LPS for 24 hours. Data are expressed as means ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001.