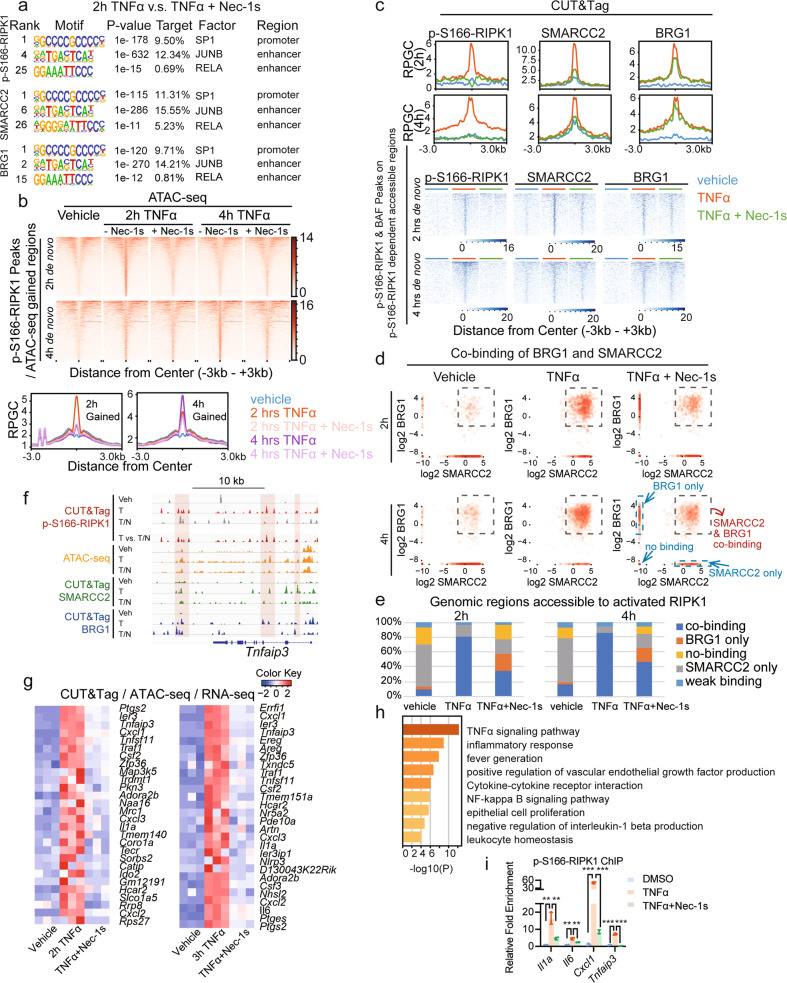
Fig. 3. Inhibition of RIPK1 kinase activity disrupts genome-wide DNA accessibility by repressing the co-occupancy of SMARCC2 and BRG1.
a HOMER motif analysis of the binding peaks of p-S166-RIPK1, SMARCC2 or BRG1 on promoters or enhancers detected by CUT&Tag under the treatment of TNFα versus TNFα plus Nec-1s. b Heatmap (top) and summary plots (below) of ATAC-seq genomic accessibility reads under the indicated conditions over the de novo sites in Ripk1D325A/D325A;Ripk3−/− MEFs treated with TNFα for 2 h or 4 h. The de novo sites were calculated by the intersection of p-S166-RIPK1-binding peaks with the accessible genomic regions induced by TNFα. c The summary plots (top) and binding intensities (below) of p-S166-RIPK1, SMARCC2 and BRG1 under the treatment of vehicle, TNFα and TNFα plus Nec-1s were shown by CUT&Tag reads over the defined sites (See Supplementary information, Fig. S3c) in Ripk1D325A/D325A;Ripk3−/− MEFs treated with TNFα for 2 h (1110 sites) or 4 h (2159 sites). The defined sites were calculated by the intersection between p-S166-RIPK1/SMARCC2/BRG1 co-binding peaks under the treatment of TNFα and RIPK1 kinase activity-dependent accessible regions. The RIPK1 kinase activity-dependent accessible regions were defined as the accessible regions that induced by TNFα and became less accessible when adding Nec-1s. d Log2-transformed binding scores of SMARCC2 and BRG1 were plotted to show the co-occupancy of SMARCC2 and BRG1 under the treatment of vehicle, TNFα and TNFα plus Nec-1s over the interested genomic regions defined in Supplementary information, Fig. S3c. Each point represents a specific site with the binding of SMARCC2 (x value) and BRG1 (y value). The sites selected by the square are defined as SMARCC2/BRG1 co-binding sites, while the sites which are plotted on x axis are defined as sites bound with SMARCC2 only, and the ones on y axis are defined as sites bound with BRG1 only. The sites which are plotted on the minus infinity of x axis and y axis are defined as no-binding sites, and the rest are defined as weak binding sites. e Summary of the percentage changes of SMARCC2/BRG1 co-binding, SMARCC2 only, BRG1 only, no-binding and weak binding sites upon the treatment of vehicle, TNFα and TNFα plus Nec-1s in d. The genomic regions used here are defined in Supplementary information, Fig. S3c. f The ATAC-seq profiles (orange) and CUT&Tag profiles of p-S166-RIPK1 (red), SMARCC2 (green) and BRG1 (blue) genomic binding upon the treatment of vehicle, TNFα and TNFα plus Nec-1s on Tnfaip3 gene locus. g Heatmap showing the scaled normalized counts in RNA-seq data of the selected genes. These genes are not only induced by TNFα and repressed by TNFα plus Nec-1s at 2 h (42 genes) or 3 h (45 genes), but also the genomic targets of p-S166-RIPK1, SMARCC2 and BRG1 (in CUT&Tag) whose accessibility is induced by TNFα and disrupted by TNFα plus Nec-1s (in ATAC-seq). See also Supplementary information, Table S2. h Pathway analysis by Metascape for the selected target genes of p-S166-RIPK1, SMARCC2 and BRG1 in g. i p-S166-RIPK1 ChIP-qPCR in Ripk1D325A/D325A;Ripk3−/− MEFs treated with vehicle, TNFα and TNFα plus Nec-1s. For each position, ChIP binding normalized to input is shown as fold change to the condition treated with vehicle. n = 3, **P < 0.01, ***P < 0.001.