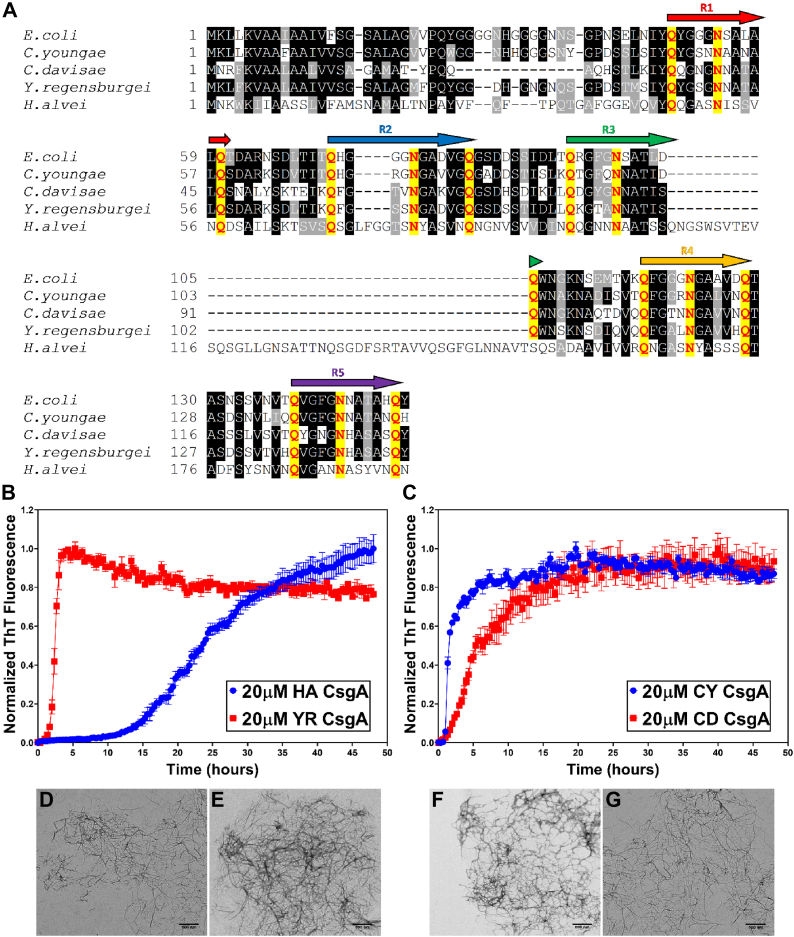
Figure 1.
Gut bacterial CsgA homologs are amyloidogenic in nature.A, sequence alignment of CsgA homologs. The five amyloid imperfect repeat units with Q-(X)4-N-(X)5-Q motif are marked as R1 to R5. The Q and N residues in the motifs are marked in red. The alignment was done using T-Coffee (http://tcoffee.crg.cat/apps/tcoffee/do:regular) with default parameters and visualized using Boxshade (https://embnet.vital-it.ch/software/BOX_form.html). Aggregation kinetics of CsgA homologs purified from the human microbiome were monitored using Thioflavin T fluorescence over the course of 48 h at 37 °C. B, 20 μM HA CsgA (blue) and 20 μM YR CsgA (red). C, 20 μM CY CsgA (blue) and 20 μM CD CsgA (red). (Error bars represent SEM of three replicates). D–G, negatively stained transmission electron micrographs of CsgA fibers. Representative images of the samples were taken 48 h postaggregation. D, HA CsgA. E, YR CsgA. F, CY CsgA. G, CD CsgA. (The scale bars represent 500 nm). CD CsgA, Cedecea davisae CsgA; CY CsgA, Citrobacter youngae CsgA; HA CsgA, Hafnia alvei CsgA; YR CsgA, Yokenella regensburgei CsgA.