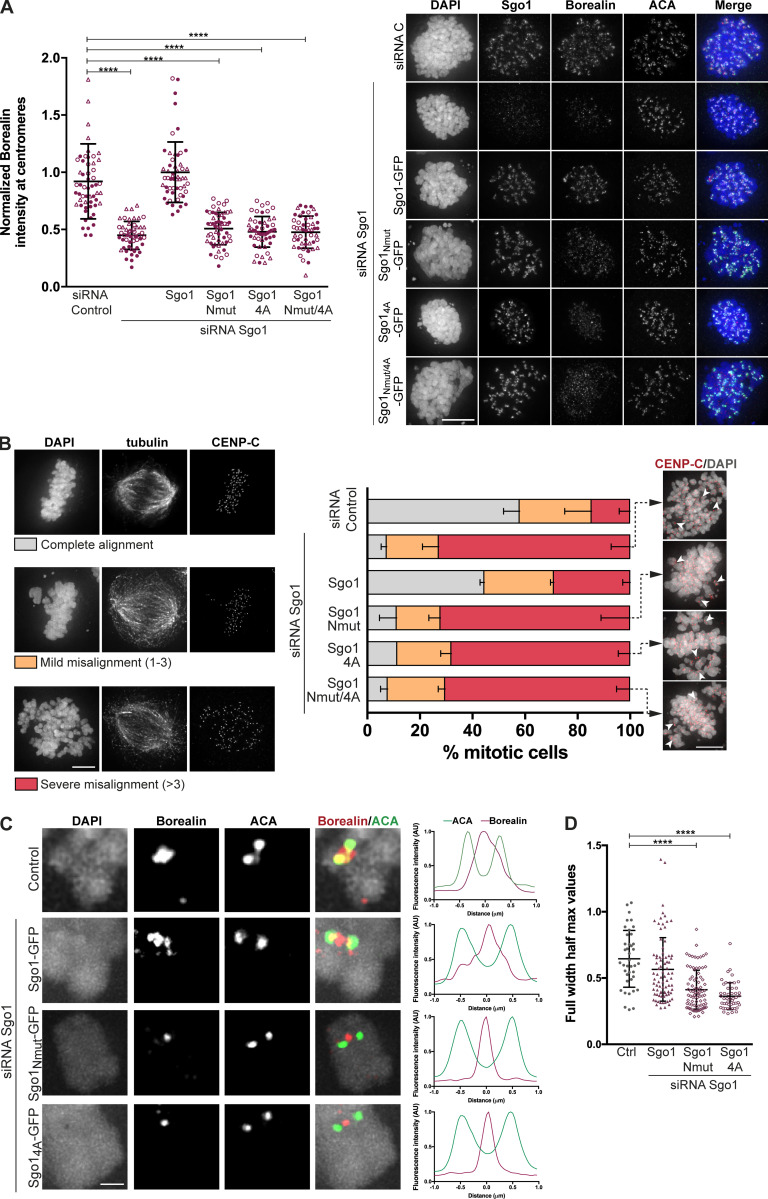
Figure 5.
CPC interaction with the Sgo1 N-terminal tail is essential for the centromeric localization of CPC and proper chromosome segregation. (A) Representative micrographs of HeLa Kyoto cells transiently expressing different Sgo1-GFP constructs (Sgo1-GFP, Sgo1Nmut-GFP, Sgo14A-GFP, or Sgo1Nmut/4A-GFP) and depleted of endogenous Sgo1 using siRNA oligonucleotides (right). Immunofluorescence of endogenous Borealin and ACA. DAPI was used for DNA staining. Scale bar, 10 µm. Quantification of Borealin levels at the centromeres using ACA as reference channel (left). Values normalized to Sgo1 siRNA/Sgo1-GFP condition. Three independent experiments, n ≥ 50 cells analyzed in total per treatment, mean ± SD, Kruskal–Wallis with Dunn’s multiple comparisons test; ****, P ≤ 0.0001. The values from the three independent replicates are represented in three different symbols. (B) Quantification of chromosome alignment of cells subjected to biorientation assay. Transfected cells were treated with 100 μM Monastrol for 16 h and released into medium containing 5 μM MG132 for 1 h. Representative examples of the alignment categories: complete alignment, mild misalignment (with one to three misaligned chromosomes), and severe misalignment (with more than three misaligned chromosomes) are found in the left panel. Representative images of the conditions expressing the three Sgo1 mutants showing pairs of CENP-C foci (red; right panel). DAPI was used to visualize DNA. Scale bar, 5 μm. Three independent experiments, n ≥ 100 of metaphases analyzed; mean ± SD. (C) Line plots depicting normalized fluorescence intensity levels of Borealin and ACA, measured along a line across the two sister ACA signals of the interkinetochore axis. Scale bar, 2 μm. Left, representative images of kinetochore pairs represented in the line plots. (D) Quantification of the full width at half maximum for the Borealin signal obtained in the line plots. Three independent experiments, n ≥ 49 kinetochores, mean ± SD, Kruskal–Wallis with Dunn’s multiple comparisons test; ****, P ≤ 0.0001.