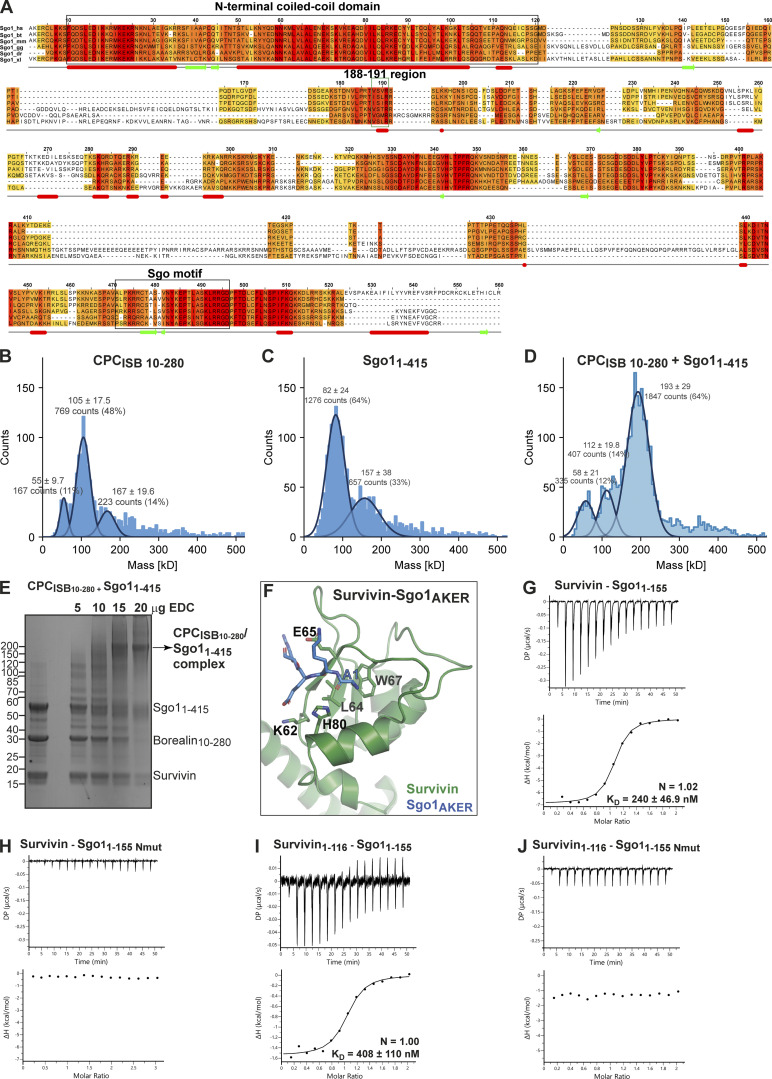
Figure S1.
CPC and Sgo1 interact in vitro. (A) Sequence alignment of Sgo1 orthologues from Homo sapiens (hs), Bos taurus (bt), Mus musculus (mm), Gallus gallus (gg), Danio rerio (dr), and Xenopus laevis (xl). The conservation score is mapped from red (highly conserved) to yellow (poorly conserved). Predicted secondary structure elements are shown below the sequence alignment. Multiple sequence alignment was performed with Clustal Omega (EMBL-EBI) and edited with Jalview 2.11.0 (Waterhouse et al., 2009). Highlighted with boxes are the N-terminal coiled-coil domain of Sgo1, the highly conserved 188–191 region, and the Sgo motif. The N-terminal AKER motif of Sgo1 is well conserved in most higher vertebrates. (B–D) Resulting mass photometry histograms and kernel density estimates for CPCISB10–280 (B), Sgo11–415 (C), and CPCISB10–280 /Sgo11–415 complex (D). All samples were cross-linked with 0.01% glutaraldehyde for 5 min at 4°C. Mean ± SD. (E) Representative SDS-PAGE analysis of CPCISB10–280 cross-linked with Sgo11–415 using EDC chemical cross-linker. (F) Close-up of the crystal structure of Survivin bound to a peptide comprising the four first amino acid residues of Sgo1 (AKER peptide; PDB accession no. 4A0I; Jeyaprakash et al., 2011). Sgo1Nmut disrupts the interaction between the first amino acid of Sgo1 and the shallow hydrophobic pocket of Survivin. Mutation of amino acids Lys62, Glu65, and His80 in the Survivin BIR domain to alanine disrupt the crucial interactions with the AKER N-terminal tail of Sgo1. (G and H) Isotherms for the analyses of Survivin interaction with Sgo11–155 (G) and Sgo11–155 Nmut (H). (I and J) Isotherms for the analyses of Survivin1–116 interaction with Sgo11–155 (I) and Sgo11–155 Nmut (J). The ITC experiments were performed with 16 × 2.5-μl injections of 200 μM Survivin or Survivin1–116 into 200 μl of 20 μM Sgo11–155 or Sgo11–155 Nmut (0.5 μl first injection), 180 s apart, at 20°C. Top panels show raw ITC data; bottom panels show integrated heat data corrected for heat of dilution and fitted to a standard 1:1 binding model (Malvern Instruments MicroCal Origin software, v1.3). DP, differential power. Source data are available for this figure: SourceData FS1.