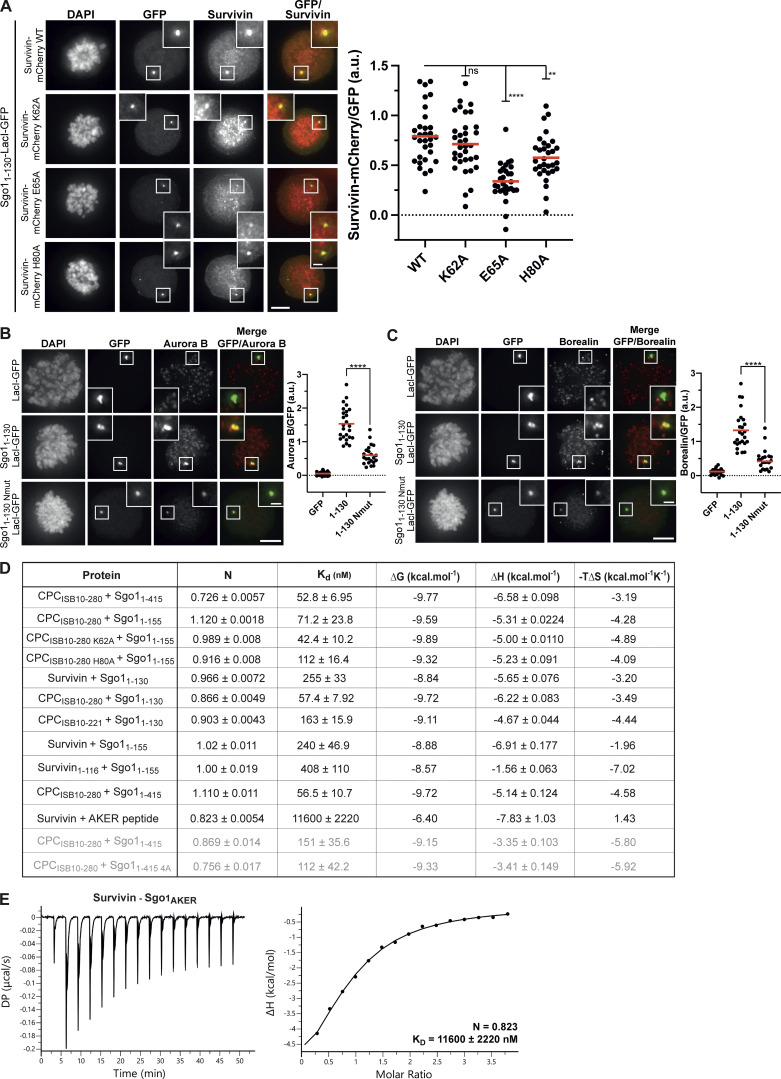
Figure S3.
Sgo1 N-terminal tail is crucial for CPC–Sgo1 interaction. (A) Representative micrographs (left) and quantifications (right) for the analysis of Survivin-mCherry WT (n = 31), K62A (n = 34), E65A (n = 29), or H80A (n = 31) recruitment to the LacO array in U-2 OS-LacO Haspin CM cells expressing Sgo11–130-LacI-GFP. One-way ANOVA with Dunnett’s multiple comparison test (****, P < 0.0001; **, P = 0.0026). Scale bar, 5 μm. (B and C) Representative micrographs (left) and quantifications (right) for the analysis of endogenous Aurora B (B) and Borealin (C) recruitment to the LacO array in U-2 OS-LacO Haspin CM expressing different Sgo1-LacI-GFP constructs: LacI-GFP (n = 21 for Aurora B; n = 21 for Borealin), Sgo11–130-LacI-GFP (n = 25 for Aurora B; n = 25 for Borealin), or Sgo11–130 Nmut-LacI-GFP (n = 22 for Aurora B; n = 22 for Borealin). Representative immunofluorescence images in B and C show Aurora B and Borealin signal for the same cell; thus, DAPI and GFP in B and C are the same. The graphs show the intensities of Survivin-mCherry, Aurora B, or Borealin over GFP (dots) and the means (red bar). Data are representative of two or five biological replicates. Scale bar, 5 μm. One-way ANOVA with Dunnett’s multiple comparison test (****, P < 0.0001). (D) Table including the ITC thermodynamic parameters for the different ITC experiments. All ITC experiments were performed using a buffer composed of 50 mM Hepes, 150 mM NaCl, 5% glycerol, and 1 mM TCEP, pH 8, except the two runs that are shown in light gray that were performed using a buffer composed of 50 mM Hepes, 250 mM NaCl, 5% glycerol, 0.005% Tween, and 1 mM TCEP, pH 8. (E) Isotherms for the analyses of Survivin interaction with Sgo1AKER peptide (40 μl of 375 μM Survivin was injected into 200 μl of 20 μM Sgo1AKER). The ITC experiment was performed with 16 × 2.5-μl injections (0.5 μl first injection), 180 s apart, at 10°C. Left panel shows raw ITC data; right panel shows integrated heat data corrected for heat of ligand dilution and fitted to a standard 1:1 binding model (Malvern Instruments MicroCal Origin software, v1.3). DP, differential power.