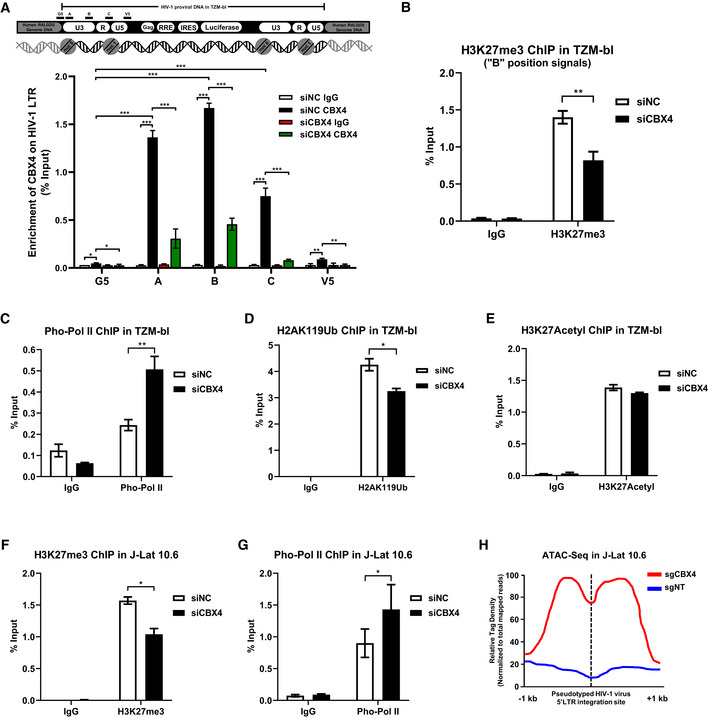
Figure 2. CBX4 contributes to H3K27me3 modification of HIV‐1 promoter.
-
AThe upper schematic represents the pseudotyped HIV‐1 provirus and corresponding integration site in TZM‐bl cells. The HIV‐1 mini‐genome was integrated in the intron of human RALGDS gene. Five pairs of ChIP‐qPCR primers were designed on the HIV‐1 LTR, which are indicated above the backbone. G5: Cellular DNA and viral 5’LTR junction; A: Nucleosome (Nuc) 0 assembly site; B: Nuc‐free region; C: Nuc 1 assembly site; V5: Viral 5’LTR and gag leader sequence junction. ChIP assays with antibodies against IgG and CBX4 were performed in both siNC‐treated and siCBX4‐treated TZM‐bl cells. All the ChIP‐qPCR DNA signals were normalized to Input. The lower statistical graph represents the ChIP‐qPCR results.
-
B–EChIP assays with antibodies against H3K27me3, Pho‐Pol II, H2AK119Ub and H3K27Acetyl were performed in TZM‐bl cells as in (A). Only “B” position signals are shown and normalized to Input.
-
F, GChIP assays with antibodies against H3K27me3 and Pho‐Pol II were performed in J‐Lat 10.6 cells as in (A). ChIP‐qPCR DNA signals on position “B” were normalized to Input.
-
HATAC‐Seq was performed in WT and CBX4‐knockout J‐Lat 10.6 cells. The relative tag densities of the pseudotyped HIV‐1 5’LTR integration site in each group were calculated. The highest tag density was set as 100. Data represent 2 kb ranges of tag densities centered the 5’LTR integration site. The integration site is indicated with dashed line.
Data information: Data are presented as mean ± SEM in biological triplicate. P‐values in (A) were calculated by two‐way ANOVA with Tukey's multiple comparisons test. P‐values in (B–G) were calculated by Student's t‐test. *P < 0.05, **P < 0.01, ***P < 0.001.
