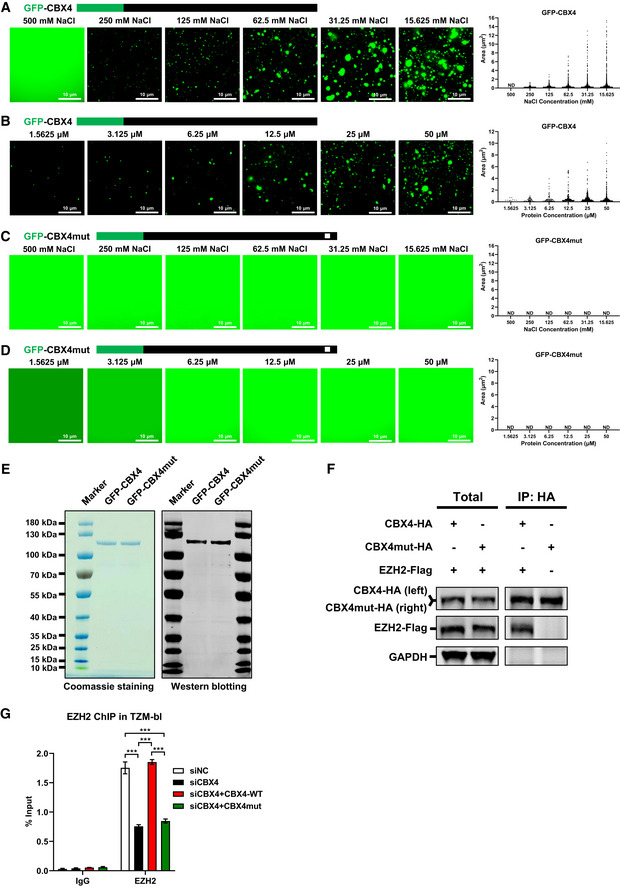
Figure EV4. The LLPS of CBX4 depends on the β‐sheet motifs of CBox.
-
AAbout 25 μM of in vitro purified GFP‐CBX4 proteins were incubated with gradient droplet formation buffers. GFP‐CBX4 droplets appeared in 250, 125, 62.5, 31.25 and 15.625 mM NaCl. The right subpanel represents statistical analysis of droplet areas and numbers of GFP‐CBX4 within different NaCl concentrations.
-
BDifferent concentrations of in vitro purified GFP‐CBX4 proteins were incubated with 75 mM NaCl droplet formation buffer. The right subpanel represents statistical analysis of droplets areas and numbers of GFP‐CBX4 within different protein concentrations.
-
C, DThe in vitro purified GFP‐CBX4mut proteins (25 μM) were incubated with different droplet formation buffers which contained different NaCl concentrations (C). Different concentrations of GFP‐CBX4mut proteins were incubated with 75 mM NaCl droplet formation buffer (D). The right subpanel within each panel represents the statistical analysis of droplets areas and numbers of GFP‐CBX4mut within different conditions.
-
EThe purities of in vitro purified GFP‐CBX4 and GFP‐CBX4mut proteins were analyzed by both Coomassie blue staining and Western blotting against GFP.
-
FHA‐tagged CBX4 and CBX4mut were co‐overexpressed with Flag‐tagged EZH2 respectively. HA‐tagged proteins were IP with anti‐HA beads. Both total and IP samples were IB with anti‐HA and anti‐Flag antibodies.
-
GTZM‐bl cells were treated with siCBX4 targeting 3’UTR of CBX4 mRNA or siNC. Another two groups of siCBX4‐treated cells were re‐overexpressed with wild‐type CBX4 and LLPS‐deficient CBX4mut respectively. ChIP assays with antibodies against IgG and EZH2 were performed in these cells.
Data information: Scale bars in (A–D) represent 10 μm. All the samples were imaged to obtain at least three sets of images. Data in (G) are presented as mean ± SEM in biological triplicate. P‐values in (G) were calculated by two‐way ANOVA with Tukey's multiple comparisons test. ***P < 0.001.
