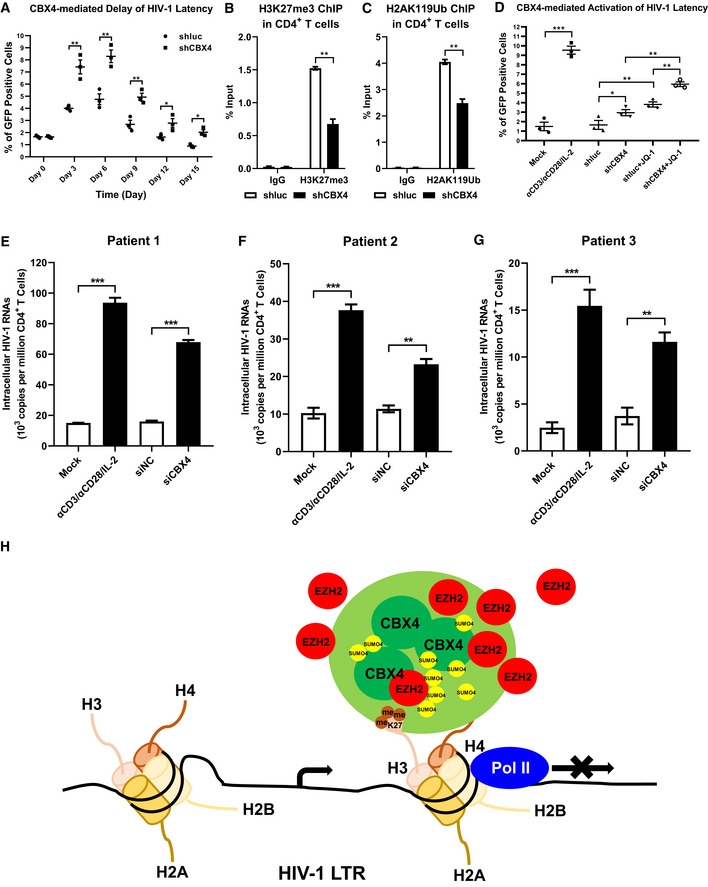
Figure 7. CBX4 depletion reactivates latent HIV‐1 in cells from HIV‐1‐infected individuals.
-
APHA‐activated primary CD4+ T cells were infected with wild‐type HIV‐1 viruses which harbored a GFP ORF after Nef gene, followed by infecting with shluc and shCBX4 lentiviruses. The percentages of GFP‐positive cells, which were HIV‐1‐expressing cells, were monitored every three days.
-
B, CPrimary CD4+ T cells were treated as in (A). On Day 6 post infection with HIV‐1 viruses and shRNA lentiviruses, ChIP assays with antibodies against H3K27me3 and H2AK119Ub were conducted in both shluc and shCBX4 groups.
-
DPHA‐activated CD4+ T cells were infected with wild‐type HIV‐1 viruses as in (A). One group of HIV‐1‐infected CD4+ T cells were left untreated. One group of HIV‐1‐infected cells were infected with shluc. Another group of HIV‐1‐infected cells were infected with shCBX4. Two weeks later, one part of HIV‐1‐infected CD4+ T cells were reactivated by αCD3/αCD28/IL‐2. One part of shluc‐ and shCBX4‐infected cells were co‐stimulated with LRA JQ‐1. GFP‐positive cells in each group were measured by flow cytometry to indicate reactivated HIV‐1‐infected cells.
-
E–GPrimary CD4+ T cells, which contained latent HIV‐1‐infected cells, were isolated from three HIV‐1‐infected individuals. One group of cells were untreated. One group of cells were activated with αCD3/αCD28/IL‐2. Another two groups of cells were transfected with siNC and siCBX4 utilizing 4D‐Nucleofector System. Three days post transfection, the amounts of intracellular HIV‐1 RNAs within each group were quantitated by RT‐qPCR and represented as 103 copies per million CD4+ T cells.
-
HThe schematic of CBX4 body‐mediated HIV‐1 latency. Detailed information for the schematic is given in the Discussion section.
Data information: Data are presented as mean ± SEM in biological triplicate. P‐values in (A–C) were calculated by two‐way ANOVA with Sidak's multiple comparisons test. P‐values in (D–G) were calculated by one‐way ANOVA with Tukey's multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001.
