Abstract
Objective
White adipose tissue (WAT) possesses the remarkable remodeling capacity, and maladaptation of this ability contributes to the development of obesity and associated comorbidities. Calsyntenin-3 (CLSTN3) is a transmembrane protein that promotes synapse development in brain. Even though this gene has been reported to be associated with adipose tissue, its role in the regulation of WAT function is unknown yet. We aim to further assess the expression pattern of CLSTN3 gene in human adipose tissue, and investigate its regulatory impact on WAT function.
Methods
In our study, we observed the expression pattern of Clstn3/CLSTN3 gene in mouse and human WAT. Genetic association study and expression quantitative trait loci analysis were combined to identify the phenotypic effect of CLSTN3 gene variant in humans. This was followed by mouse experiments using adeno-associated virus-mediated human CLSTN3 overexpression in inguinal WAT. We investigated the effect of CLSTN3 on WAT function and overall metabolic homeostasis, as well as the possible underlying molecular mechanism.
Results
We observed that CLSTN3 gene was routinely expressed in human WAT and predominantly enriched in adipocyte fraction. Furthermore, we identified that the variant rs7296261 in the CLSTN3 locus was associated with a high risk of obesity, and its risk allele was linked to an increase in CLSTN3 expression in human WAT. Overexpression of CLSTN3 in inguinal WAT of mice resulted in diet-induced local dysfunctional expansion, liver steatosis, and systemic metabolic deficiency. In vivo and ex vivo lipolysis assays demonstrated that CLSTN3 overexpression attenuated catecholamine-stimulated lipolysis. Mechanistically, CLSTN3 could interact with amyloid precursor protein (APP) in WAT and increase APP accumulation in mitochondria, which in turn impaired adipose mitochondrial function and promoted obesity.
Conclusion
Taken together, we provide the evidence for a novel role of CLSTN3 in modulating WAT function, thereby reinforcing the fact that targeting CLSTN3 may be a potential approach for the treatment of obesity and associated metabolic diseases.
Keywords: Calsyntenin-3, Obesity, Dysfunctional adipose tissue, Amyloid precursor protein, Adipose mitochondrial dysfunction
Highlights
-
•
CLSTN3 is expressed in the adipocyte fraction of human adipose tissue and mainly localizes to the plasma membrane.
-
•
SNP rs7296261 in human CLSTN3 locus is associated with obesity risk.
-
•
Overexpression of CLSTN3 leads to adipose tissue dysfunction in mice.
-
•
CLSTN3 can attenuate catecholamine-stimulated lipolysis.
-
•
CLSTN3 overexpression increases mitochondrial APP localization of mouse adipose tissue.
1. Introduction
Understanding the processes that lead to excess adiposity is necessary to elucidate the pathophysiology of obesity. It facilitates to identify novel avenues for preventing and treating obesity-related disorders [1]. In adults, an increase in adipocyte number (hyperplasia) may lead to gain of lower-body fat, while an increase in adipocyte size (hypertrophy) may cause expansion of abdominal white adipose tissue (WAT) [2]. Adipocyte hypertrophy contributes to an increased risk of hypoxia, which consistently induces stress signaling that initiates chronic low-grade inflammation and triggers fibrotic program in WAT. The integrated response promotes adipocyte dysfunction as well as whole-body metabolic defects [3,4]. Moreover, increased subcutaneous adipocyte size, particularly in the abdominal region, is a predictor of obesity-associated comorbidities, such as type 2 diabetes [5]. Accordingly, targeting adipocyte hypertrophy may help in improving metabolic dysfunction in people with obesity. However, the factors driving adipocyte enlargement and fat accumulation have not yet been completely identified.
Despite the presence of the environmental factors that drive fat accumulation, a twin study clearly demonstrated a substantial degree of genetic control on human adiposity [6]. Genome-wide association study (GWAS), a high-throughput genotyping technology, has remarkably increased the speed of gene discovery at loci with associations for common traits and diseases, including obesity [[7], [8], [9]]. The fat mass and obesity-associated (FTO) gene is the first obesity susceptibility gene to be identified by GWAS, and this locus has the largest effect on body mass index (BMI) and obesity risk [10,11]. Furthermore, genome-wide polygenic scores integrate all available variants to quantify the inherited susceptibility to obesity from birth to adulthood [12]. A list of candidate genes has been discovered that may be responsible for the development of obesity in different populations [13]. Subsequent functional studies of these genes have elucidated the biological pathways involved in adipose biology [14,15]. However, there is still a great scope of discovering obesity-associated genes using human genomic datasets and analyzing their functional relevance with fat accumulation in the context of obesity.
Calsyntenins (CLSTNs) are evolutionarily conserved proteins that were originally identified in central nervous system (CNS), and play critical roles during neural development [16,17]. Calsyntenin-3 (CLSTN3) is localized to the postsynaptic membrane where it acts as a synaptogenic adhesion molecule via interaction with Neurexin 1α [18]. However, few studies have investigated the roles of CLSTN3 in peripheral tissues of mice and humans. Amyloid precursor protein (APP) is also a transmembrane protein that is widely expressed in CNS as well as peripheral tissues, including liver and adipose tissue. Abnormal expression of APP in peripheral tissues is associated with metabolic diseases, such as type 2 diabetes and nonalcoholic fatty liver disease [19,20]. Interestingly, CLSTN3 can interact with APP and the neural adaptor protein X11-like to form a tripartite complex in brain that enhances the stabilization of APP metabolism [21,22]. The similar expression pattern of CLSTN3 and APP in CNS as well as the interaction between them suggest that they may share common or coordinated regulatory roles in peripheral tissues.
In this study, the transcript of Clstn3/CLSTN3 gene was observed from transcriptomic data of WAT in mice and humans, respectively. Thereafter, we explored the phenotypic effect of CLSTN3 gene variant using human genetic association study and expression quantitative trait loci (eQTL) analysis. We proposed that obesity risk conferred by CLSTN3 rs7296261 is mediated by high CLSTN3 expression in human adipose tissue. Additionally, we aimed to assess the effect of CLSTN3 overexpression in inguinal WAT (iWAT) of mice on adipose tissue function, and determine it as a potential target for preventing and treating obesity and associated comorbidities.
2. Materials and methods
2.1. Subjects
A total of 2,386 individuals were recruited from Shanghai Obesity Study (SHOS) [[23], [24], [25]]. Detailed study methods about recruitment and clinical data collection have been previously described [23]. Briefly, SHOS is prospective cohort to investigate the development of obesity and associated diseases. Genomic DNA was isolated from whole blood sample and then subjected to exome genotyping. The associations of the single nucleotide polymorphisms (SNPs) in the CLSTN3 locus with obesity-associated traits were obtained. These individuals were grouped by SNP rs7296261 and the clinical characteristics were presented in Table S1. The eQTL analysis for CLSTN3 rs7296261 on gene expression was performed in paired abdominal subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT) from 81 obese participants who underwent bariatric surgery at Shanghai Jiao Tong University Affiliated Sixth People's Hospital. Patients with severe conditions were not included, including generalized inflammation or malignant diseases. The clinical characteristics of these participants grouped by CLSTN3 rs7296261 genotyping were presented in Table S2. Lastly, we analyzed the abdominal SAT from 10 lean (BMI, 17.9–24.4 kg/m2) and 10 obese (BMI, 31.1–42.3 kg/m2) participants, who underwent a laparoscopic cholecystectomy or bariatric surgery at Shanghai Jiao Tong University Affiliated Sixth People's Hospital. All human studies were approved by the Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital. All participants provided written informed consent.
2.2. Genotyping and quality control
Genomic DNA was extracted from peripheral blood leukocytes using QIAamp DNA Blood Mini Kit (Qiagen), according to the manufacturer's instructions. Genome-wide genotyping and quality control of extracted DNA were performed, according to previously published protocols [24]. In brief, genome-wide genotyping of all the subjects was performed using Infinium Exome-24 v1.0 BeadChip (Illumina). DNA quality control was evaluated at the individual as well as the SNP level. We screened three variants in the CLSTN3 locus, including rs145190321 (MAF = 0.0002), rs189282788 (MAF = 0.0027), and rs7296261 (MAF = 0.4765). The first two loci were not included due to the low-frequency in the population. CLSTN3 rs7296261 was further genotyped in genomic DNA samples of 81 obese participants who underwent bariatric surgery by sanger sequencing following polymerase chain reaction (PCR) amplification. For quality control, resequencing validation of randomly chosen individuals was done to confirm genotyping results.
2.3. Animals
Male C57BL/6 mice, aged 4–5 weeks, were purchased from Shanghai SLAC Laboratory Animal Company and housed under a 12-hour light/dark cycle with free access to water and food. Adeno-associated virus (AAV) was administered into iWAT pads of 6-week-old mice. To induce WAT browning model, 9-week-old mice were individually caged at 4 °C for 1 week. To induce obesity model, 7-week-old mice were fed on high-fat diet (HFD) for 12–15 weeks, in which 60% kcal was obtained from fat, 20% kcal from carbohydrate and 20% kcal from protein (Research Diets, D12492). For cold-induced lipolysis, mice were exposed to 4-hour cold stress. For in vivo lipolysis, β3-adrenoceptor agonist CL-316,243 (MilliporeSigma) was intraperitoneally injected to the mice at a dose of 1 mg/kg body weight. All animal studies were reviewed and approved by the Animal Care Committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital.
2.4. Production and injection of AAV
The coding sequence of human CLSTN3 transcript (GenBank, NM_014718.4) was a generous gift from Jiahuai Han's laboratory (School of Life Sciences, Xiamen University, China). It was cloned into pAAV-CMV-3FLAG-WPRE vector (Obio Technology, Shanghai, China) to induce overexpression of CLSTN3 (AAV-CLSTN3, Figure S1A). A viral vector without CLSTN3 plasmid insertion was used as the control (AAV-CON, Figure S1A). The full sequence map for pAAV-CMV-CLSTN3-3FLAG-WPRE vector and CLSTN3 coding sequence was shown in Figure S1B and Figure S1C, respectively. The vectors were packaged into AAV9 (GenBank, AY530579.1) by Obio Technology. Approximately 4 × 1010 AAV particles were administered into both sides of iWAT pads of each mouse. After 4 weeks, mice were sacrificed to examine the efficiency of AAV-mediated CLSTN3 overexpression in iWAT.
2.5. Human primary adipocytes
Stromal vascular fraction (SVF) and mature adipocytes were isolated from human abdominal SAT as previously described [26]. Fresh adipose tissue was collected in Dulbecco's Modified Eagle Medium (DMEM, MilliporeSigma) and kept on ice during the transport. Biopsy was minced and digested with DMEM containing 0.2% type II collagenase (MilliporeSigma) and 1.5% bovine serum albumin (BSA) for 30 min at 37 °C with gentle shaking. After being kept on ice for 10 min, digested sample was filtered with a 70 μm cell strainer, and cell suspension was centrifuged for 10 min at 800×g. The cell precipitate was washed with phosphate buffer saline (PBS) and centrifuged again to get SVF. The floating mature adipocyte fraction was further washed with PBS and collected for further use.
2.6. Extraction of membrane and cytosol protein
Membrane and cytosolic protein fractions were extracted from human primary adipocytes using the Membrane and Cytosol Protein Extraction Kit (Beyotime) according to the manufacturer's protocol [27]. After efficient homogenization, sample was centrifuged at 600×g to remove the nuclei and intact cells. Then, the supernatant was centrifuged at 14,000×g for 30 min at 4 °C to obtain the precipitate that was comprised of membrane fraction. Membrane protein was extracted with specific reagent. The remaining supernatant was used to obtain cytosol protein.
2.7. RNA extraction and gene expression analysis
Total RNA from human and mouse tissue was extracted with TRIzol reagent (Invitrogen). RNA concentration and purity was measured by NanoDrop (Thermo Fisher Scientific). Then, 1 μg of RNA was used to synthesize cDNA using GoScript™ Reverse Transcription System (Promega). Real-time quantitative PCR was conducted using SYBR Green Supermix (Vazyme) in a LightCycler480 PCR system (Roche). Quantitative levels of gene expression were normalized to the housekeeping gene, human RPLP0 or mouse 36b4, using the 2ˆ(−ΔΔCT) method. Primers were listed in Table S3.
2.8. Immunoblot analysis
Tissue was homogenized, and protein was extracted using radioimmunoprecipitation lysis buffer (Beyotime) containing phosphatase/protease inhibitor cocktail (Roche). Protein concentration was measured using the BCA protein assay kit (Beyotime). Equivalent amount of protein was loaded onto a sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred onto a nitrocellulose membrane (MilliporeSigma). The membrane was blocked with 5% skim milk for 1 h at room temperature, and incubated overnight at 4 °C with primary antibody against CLSTN3 (Proteintech, 13302-1-AP), CAV1 (Proteintech, 66067-1-Ig), Actin (Abcam, ab8227), Tubulin (MilliporeSigma, T6199), UCP1 (Abcam, ab10983), P-AKTThr308 (Cell Signaling Technology, 13,038), T-AKT, (Cell Signaling Technology, 9272), P-HSLSer563 (Cell Signaling Technology, 4139), T-HSL (Cell Signaling Technology, 4107), FLAG (GenScript, AF519), HA (Cell Signaling Technology, 3724), APP (GeneTex, GTX101336), VDAC1 (Abcam, ab154856), and OXPHOS rodent WB cocktail (Abcam, ab110413). Then, the membrane was incubated with secondary anti-rabbit or anti-mouse antibody for 1 h at room temperature. Protein bands were detected with ECL chemiluminescent reagent (MilliporeSigma) using the Image Quant LAS4000 System (GE Healthcare).
2.9. Histology analysis
Adipose tissue or liver sample was fixed in 4% paraformaldehyde for 24 h at 4 °C, embedded in paraffin, and cut into 4-μm thick section for histological analysis. Section was stained with hematoxylin and eosin (H&E) staining to determine morphological change, and image was viewed and photographed using a PANNORAMIC Digital Slide Scanner (2DHISTECH). Image-Pro Plus 6.0 was used to calculate adipocyte area and number from the sections of six individual mice and two fields per mouse in each group. In addition, some of histological images of were acquired with a Nikon microscope, and adipocyte area was quantified from the sections of three mice. For immunofluorescence analysis, deparaffinized tissue section was blocked with goat serum, and incubated with anti-CLSTN3 antibody (Proteintech), followed by Alexa Fluor 488-labeled Goat Anti-Rabbit IgG (Beyotime) and DAPI staining solution. Image was captured using a Leica fluorescence microscope.
2.10. Insulin signaling analysis
Insulin (Lily) at a concentration of 1.0 U/kg body weight was intraperitoneally injected to HFD-fed mice following 6-hour fasting period. Mice were sacrificed 15 min after insulin injection and iWAT pads were collected for further measurement.
2.11. Systemic metabolic tests
Glucose tolerance test (GTT) and insulin tolerance test (ITT) was performed in mice fed with HFD for 13 week and 14 weeks, respectively. For GTT, mice were subjected to overnight fasting followed by intraperitoneal injection of d-glucose solution (MilliporeSigma) at a dose of 1.25 g/kg body weight. For ITT, insulin (Lily) at a concentration of 1.0 U/kg body weight was intraperitoneally administered to the mice after a 6-hour fasting period. Blood glucose level was measured with a glucometer (Roche) at each time point, including before and 15, 30, 60, 90, and 120 min after glucose or insulin challenge.
2.12. Serum and liver chemistry
After mice were euthanized, blood samples were obtained and centrifuged to collect the serum. Serum insulin level was measured using a commercially available enzyme-linked immunosorbent assay (ELISA) kit (ImmunoDiagnostics Limited). Serum level of total cholesterol and triglyceride was measured using commercially available kits from Siemens Healthcare Diagnostics Inc. with ADVIA 2400 Chemistry System [28]. Triglyceride in liver was determined with Triglyceride Quantification Colorimetric/Fluorometric Kit (Biovision) following the manufacturer's protocol [29]. Briefly, after homogenizing approximately 50 mg of liver tissue, the lysate was centrifuged to obtain the supernatant for hepatic triglyceride content and total protein concentration measurements.
2.13. Lipolysis assay
For cold-induced and in vivo lipolysis, blood samples were collected at different time points, and centrifuged at 4,000 rpm for 10 min to get mouse serum. For ex vivo lipolysis, approximately 20 mg of iWAT explant was weighed, placed into a 24-well plate with 200 μl of DMEM containing 2% BSA, and incubated with 10 μmol/L isoproterenol (MilliporeSigma), a β-adrenoceptor agonist, at 37 °C incubator for 60 min. Medium was collected at different time intervals for further measurement. The level of free glycerol and non-esterified fatty acid (NEFA) in mouse serum or culture medium was measured using commercially kits, respectively (Free glycerol, MilliporeSigma; NEFA, Wako).
2.14. Immunoprecipitation assay
Immunoprecipitation assay was performed according to a previously described protocol [30]. For in vitro immunoprecipitation, CLSTN3-FLAG and APP-HA plasmids were co-transfected into 293T cells (ATCC) in 6-well plate using Lipofectamine (Invitrogen) following the manufacturer's instructions. Cells were harvested after 48 h later, and resuspended in lysis buffer (Beyotime) containing protease inhibitor (Roche). For FLAG-tagged immunoprecipitation in mice, iWAT samples were homogenized with lysis buffer. The lysate from cultured cells or tissues was centrifuged with 12,000 rpm for 10 min at 4 °C. Supernatant was harvested and incubated with FLAG beads (MilliporeSigma) for 4 h at 4 °C. For endogenous immunoprecipitation in human SAT, supernatant from homogenized lysate was incubated with anti-CLSTN3 antibody (Proteintech) or IgG (Beyotime) pre-treated bead (MedChemExpress) overnight at 4 °C. All the samples were analyzed by immunoblotting with indicated antibodies.
2.15. Mitochondrial isolation
Mitochondria were isolated from iWAT biopsy using Tissue Mitochondria Isolation Kit (Beyotime, C3606). After mice were euthanized, iWAT pads were removed rapidly and minced into small pieces. A glass homogenizer was used to grind the tissue pieces on ice with mitochondria isolation solution containing protease inhibitor (Roche). The lysate was centrifuged with 600×g for 5 min at 4 °C, and supernatant was further centrifuged with 11,000×g for 10 min at 4 °C. The precipitate containing mitochondria was resuspended with mitochondrial lysate solution. Mitochondrial protein concentration was measured by the BCA method.
2.16. Mitochondrial respiration measurement
Ex vivo mitochondrial respiration was measured using Seahorse XF24 Extracellular Flux Analyzer (Agilent) as previously described [31]. Firstly, 5–10 mg of iWAT explants were placed into the XF24 Islet Capture Microplates (Agilent), and incubated with XF base medium with 25 mmol/L glucose, 2 mmol/L glutamine, and 2 mmol/L sodium pyruvate (pH 7.4) in a 37 °C non-CO2 incubator (Agilent) for 1 h. Then, tissue explants were sequentially injected with 10 μmol/L oligomycin, 8 μmol/L FCCP, and 12 μmol/L antimycin A plus rotenone to perform mitochondrial stress test. The initial oxygen consumption rate (OCR) value was assessed by Seahorse XF24 software, and the final OCR result was standardized to the protein content of tissue explant in each well. The primary data of initial OCR value and protein content were annotated in Table S4.
2.17. Mitochondrial DNA copy number
Mitochondrial DNA (mtDNA) copy number was measured according to a previously described protocol [32]. Genomic DNA was extracted from mouse iWAT using QIAamp Fast DNA Tissue Kit (Qiagen). The ratio of mitochondrial gene mt–Nd1 to nuclear gene Rbm15 was determined by performing quantitative PCR. The primer sequences were listed in Table S3.
2.18. Statistical analysis
We used GraphPad Prism 7.0 and SAS 9.4 software to perform the statistical analyses. All data were presented as mean ± standard error of the mean (SEM), mean ± standard deviation (SD) or median (interquartile range, 25–75%). The two-tailed paired or unpaired Student's t test, and Wilcoxon signed rank sum test were used for establishing the comparison between two groups. The two-way ANOVA followed with Bonferroni correction was applied for multiple comparisons with two independent factors. Linear regression analysis was performed to examine the correlation between two variables. Statistical significance was set at p-value < 0.05.
3. Results
3.1. CLSTN3 gene is routinely expressed in human adipose tissue
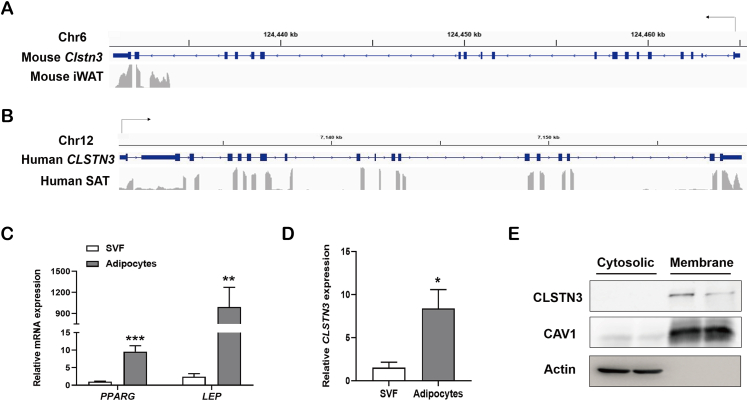
To identify the expression pattern of Clstn3/CLSTN3 gene in WAT, we analyzed the distribution of RNA sequencing reads in the Clstn3/CLSTN3 locus in iWAT of mice and SAT of humans, respectively. Integrative Genomics Viewer demonstrated that the transcript of Clstn3 gene was expressed in a very low level in iWAT of mice; it was mainly expressed in the novel form of Clstn3b gene that shares the last two exons with mouse Clstn3 gene [33] (Figure 1A). On the contrary, we observed that the transcript of CLSTN3 gene was routinely expressed in human SAT (Figure 1B). To understand the expression of CLSTN3 in human adipose tissue further, we attempted to isolate SVF and adipocyte fraction from SAT biopsies. Interestingly, mRNA expression of CLSTN3 was predominantly enriched in adipocyte fraction, which was consistent with adipocyte marker gene PPARG and LEP (Figure 1C,D). CLSTN3 acts as a synaptogenic protein that is abundantly localized at the cell surface. Here, cell membrane localization of CLSTN3 in human primary adipocytes was further supported by the evidence from subcellular fractionation and immunoblot analysis; it was identical to the membrane marker CAV1 (Figure 1E). Together, the discrepant expression pattern of Clstn3/CLSTN3 gene in mice and humans, suggests that CLSTN3 may play a specific role in human adipose biology.
Figure 1.
Identification of CLSTN3 expression in human adipose tissue. (A and B) Distribution of RNA sequencing reads in the Clstn3 locus from inguinal white adipose tissue (iWAT) of mice (A), and in the CLSTN3 locus from subcutaneous white adipose tissue (SAT) of humans (B). (C) Relative mRNA expression of adipocyte marker gene PPARG and LEP assessed by quantitative PCR in stromal vascular fraction (SVF) and mature adipocyte fraction isolated from human SAT biopsies (n = 5). (D) CLSTN3 mRNA expression in SVF and adipocytes isolated from human SAT biopsies (n = 5). (E) Immunoblot analysis of CLSTN3 protein expression in membrane fraction and cytosolic fraction isolated from human adipocytes; CAV1, membrane protein marker; Actin, cytosolic protein marker. Data are presented as mean ± standard error of the mean (SEM), and ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
3.2. The variant rs7296261 in human CLSTN3 locus is associated with obesity risk
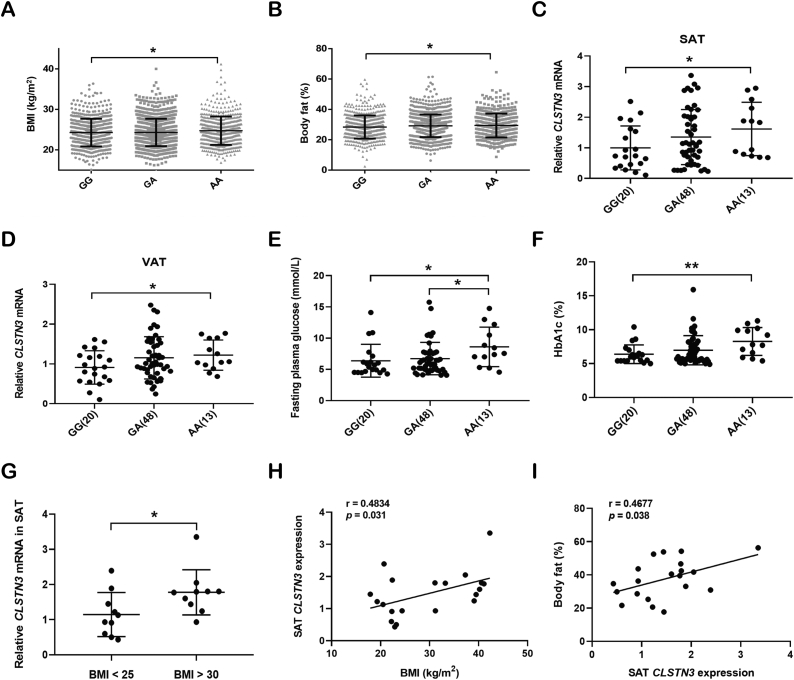
To test this hypothesis, we summarized the associations of CLSTN3 variants with obesity risk in a human cohort of 2,386 individuals from SHOS. We found that only SNP rs7296261 in the CLSTN3 locus was significantly associated with metabolic traits. Genotyping for rs7296261 divided the individuals into three groups, namely 659 GG, 1,180 GA, and 547 AA genotype carriers. The clinical characteristics of three groups were presented in Table S1. Of note, there were significant differences in metabolic traits between the homozygous GG and AA genotype carriers, including BMI, body fat, total cholesterol, and low-density lipoprotein cholesterol (LDL-c). In particular, compared to GG genotype carriers, AA genotype carriers exhibited a higher BMI (mean, 24.28 kg/m2 vs 24.74 kg/m2) and body fat (mean, 28.4% vs. 29.4%) (Figure 2A,B). Since individuals with modest BMIs in the SHOS cohort had the compensatory ability of insulin action under glucose challenge, there was no significant difference in glucose metabolism between two groups (Table S1) [34].
Figure 2.
Association of SNP rs7296261 in the CLSTN3 locus with human obesity risk. (A and B) Association between rs7296261 in the CLSTN3 locus and body mass index (BMI) (A) and body fat (B) in 2,386 individuals from Shanghai obesity study (SHOS). Individuals were grouped according to the genotyping for rs7296261, including GG (n = 659), GA (n = 1,180), and AA (n = 547) genotype carriers. (C and D) Expression quantitative trait loci (eQTL) analysis of SNP rs7296261 in the CLSTN3 locus, and CLSTN3 mRNA expression in abdominal SAT (C) and visceral adipose tissue (VAT) (D) from 81 obese participants, grouped by GG (n = 20), GA (n = 48) and AA (n = 13) genotypes. (E and F) Association between CLSTN3 rs7296261 and fasting plasma glucose (E) and HbA1c (F) in 81 obese participants. (G) CLSTN3 mRNA expression in abdominal SAT from humans with BMI <25 (n = 10) and BMI >30 (n = 10). (H and I) Correlation of CLSTN3 mRNA expression in SAT with BMI (H) and body fat (I). Values are expressed as mean ± standard deviation (SD), and ∗p < 0.05, ∗∗p < 0.01.
To further understand the relationship between CLSTN3 rs7296261 and increased adiposity, we performed eQTL analysis on paired abdominal SAT and VAT of 81 obese participants. SNP rs7296261 is located in the intronic region of CLSTN3, and the presence of this variant may alter the transcriptional expression of this gene [35]. Genotype-Tissue Expression (GTEx) database showed the eQTL analysis for rs7296261 [36]. Individuals carrying AA genotype exhibited higher CLSTN3 expression in both subcutaneous and visceral fat (Figure S2A and S2B). Our data confirmed that CLSTN3 mRNA level was higher in both SAT and VAT of individuals carrying AA genotype than that in GG genotype carriers (Figure 2C,D). SNP rs7296261 is also located in the first exon of novel CLSTN3B gene, but there was no difference in CLSTN3B mRNA level between GG and AA groups (Figure S2C and S2D). Meanwhile, we performed an additional analysis on metabolic traits of 81 obese participants with severe BMIs according to the genotyping of CLSTN3 rs7296261. Surprisingly, we observed that fasting plasma glucose (mean, 6.38 mmol/L vs 8.6 mmol/L) and HbA1c (mean, 6.39% vs 8.27%) were higher in AA genotype carriers than in GG genotype carriers (Figure 2E,F); 2-hour plasma glucose after 75 g oral glucose tolerance test (OGTT) showed an increasing trend in AA genotype group, compared to GG genotype group (mean, 9.14 mmol/L vs 11.97 mmol/L, p = 0.071) (Table S2).
Furthermore, we observed that CLSTN3 mRNA level in SAT of obese subjects was significantly elevated compared to that in lean participants (Figure 2G). SAT CLSTN3 expression was positively correlated with BMI (r = 0.4834, p = 0.031) and body fat (r = 0.4677, p = 0.038) (Figure 2H,I). Together, these results demonstrate that CLSTN3 rs7296261, which is associated with obesity risk, leads to an increase in CLSTN3 expression in human adipose tissue; moreover, high CLSTN3 expression may be associated with increased adiposity.
3.3. CLSTN3 has no effect on adipose thermogenesis in mice
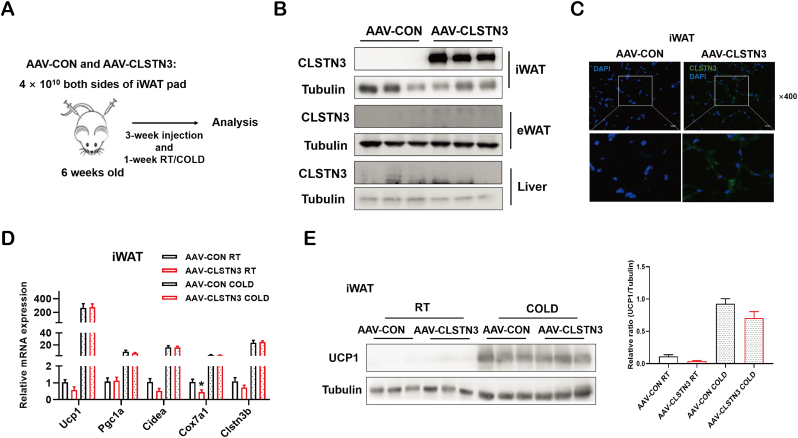
To test the hypothesis that CLSTN3 may play a role in the regulation of adipose biology in mice, we first generated an AAV carrying human CLSTN3 coding sequence (AAV-CLSTN3) and the control virus (AAV-CON). AAV particles were subcutaneously injected to both sides of iWAT pads of C57BL/6J male mice (Figure 3A). Immunoblotting and immunofluorescence analyses confirmed that CLSTN3 was locally overexpressed in iWAT; meanwhile, we did not observe the presence of endogenous Clstn3 protein expression in iWAT, epididymal WAT (eWAT) and liver (Figure 3B,C). The novel form of Clstn3 gene, Clstn3b, regulates whole-body metabolism by controlling adipose thermogenesis in mice [33]. Therefore, we examined the effect of CLSTN3 overexpression in the regulation of adipose thermogenesis. However, there was no apparent difference between AAV-CON and AAV-CLSTN3 mice in terms of the expression of thermogenic genes (Ucp1, Pgc1a, Cidea, Cox7a1, and Clstn3b) in iWAT (Figure 3D), irrespective of whether they were exposed to room temperature or chronic cold. Incidentally, UCP1 protein level remained unaltered in iWAT upon CLSTN3 overexpression (Figure 3E).
Figure 3.
CLSTN3 has no effect on iWAT browning in mice. (A) Schematic illustration of AAV injection and experimental procedures in iWAT from a mouse model. (B) Validation of CLSTN3 protein overexpression in iWAT, but not in eWAT and liver of AAV-CLSTN3 mice after 4-week AAV injection. (C) Representative images of anti-CLSTN3 (green) and DAPI (blue) immunofluorescence staining. Scale bar, 50 μm. (D) Relative mRNA levels of thermogenic genes (Ucp1, Pgc1a, Cidea, Cox7a1, and Clstn3b) assessed by quantitative PCR in iWAT of AAV-CON and AAV-CLSTN3 mice under room temperature (RT) (n = 5) and cold exposure (COLD) (n = 10) for 1 week. (E) Immunoblot analysis of UCP1 protein expression and its quantification in iWAT of AAV-CON and AAV-CLSTN3 mice under RT and COLD. Data are presented as mean ± SEM of biological independent samples, and ∗p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Overexpression of CLSTN3 causes diet-induced dysfunctional iWAT and liver steatosis
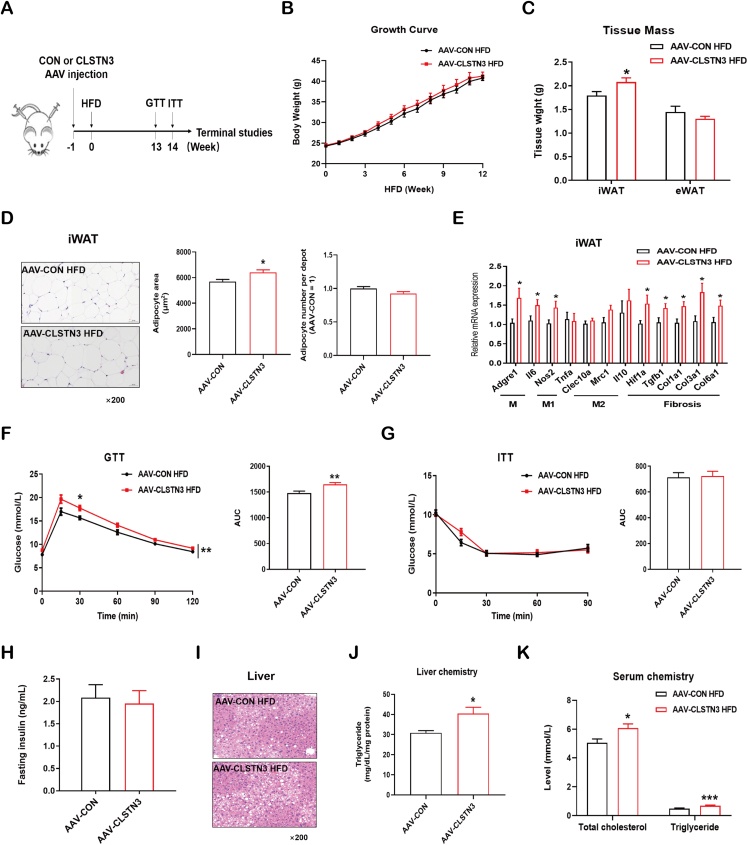
To understand whether CLSTN3 overexpression in iWAT drives diet-induced obesity, we subjected the mice to HFD feeding (Figure 4A). During 12-week HFD course, body weight of AAV-CLSTN3 mice remained similar to that of AAV-CON mice (Figure 4B). Furthermore, food intake appeared comparable in the two groups (Figure S3A). However, tissue weight of iWAT, where CLSTN3 was overexpressed, increased significantly; tissue weight of eWAT, where CLSTN3 was not overexpressed, did not exhibit a significant change (Figure 4C). Moreover, H&E staining revealed the presence of larger adipocytes without affecting the adipocyte number in iWAT of AAV-CLSTN3 mice as compared to that in AAV-CON mice (Figure 4D); eWAT adipocytes did not differ in size and number between two groups (Figure S3B). Gene expression analysis showed an increase in the expression of generic macrophage genes (Adgre1 and Il6) and pro-inflammatory M1 macrophage marker (Nos2) in iWAT of AAV-CLSTN3 mice (Figure 4E). Obesity is frequently associated with chronic adipose inflammation, which is also linked to adipose fibrosis [4]. Hence, we found that the expression of the major driver for adipose fibrosis, Hif1a, was upregulated in CLSTN3-overexpressed iWAT. This was further highlighted by increased expression of its target genes, including Tgfb1, Col1a1, Col3a1, and Col6a1 (Figure 4E). Furthermore, AAV-CLSTN3 mice showed an attenuated P-AKT signal in iWAT upon insulin stimulation (Figure S3C). The expression of genes (Slc2a1, Slc2a4, and Hk2) involved in glucose clearance showed a reducing trend in CLSTN3-overexpressed iWAT (Figure S3D). In conclusion, these data demonstrate that iWAT of AAV-CLSTN3 mice displays a local dysfunctional expansion phenomenon, which may further pose a risk for systemic health in mice.
Figure 4.
CLSTN3 overexpression deteriorates diet-induced iWAT dysfunction and liver steatosis. (A) Schematic representation of AAV injection in iWAT, and experimental procedures for AAV-CON and AAV-CLSTN3 mice fed a high-fat diet (HFD) for 12–15 weeks. (B) Growth curve of body weight during 12-week HFD (n = 8–9). (C) Tissue mass of iWAT and epididymal white adipose tissue (eWAT) at 15-week HFD (n = 7–9). (D) Representative hematoxylin and eosin (H&E) staining images of iWAT sections at 15-week HFD, and its quantification of adipocyte size and number. Scale bar, 50 μm. (E) Quantitative PCR analysis of mRNA expression of inflammation (generic macrophage signature gene: Adgre1 and Il6; M1-like: Nos2 and Tnfa; M2-like: Clec10a, Mrc1, and Il10) and fibrosis (Hif1a, Tgfb1, Col1a1, Col3a1, and Col6a1) related genes in iWAT from AAV-CON and AAV-CLSTN3 mice upon 15-week HFD (n = 9). (F) Glucose tolerance test (GTT) performed in AAV-CON and AAV-CLSTN3 mice after 13-week HFD, and its area under the curve (AUC) (n = 7–8). (G) Insulin tolerance test (ITT) performed upon 14-week HFD, and its AUC (n = 6–9). (H) Fasting insulin concentration measured in serum at 15-week HFD (n = 8–9). (I) Representative H&E staining images of liver sections at 15 weeks of HFD. Scale bar, 50 μm. (J) Triglyceride levels in liver from AAV-CON and AAV-CLSTN3 groups (n = 9–10). (K) Total cholesterol and triglyceride levels in serum from AAV-CON and AAV-CLSTN3 mice fed with 15-week HFD (n = 9–10). Data are shown as mean ± SEM of biologically independent samples, and ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Thereafter, we attempted to correlate the relevance of this phenomenon to whole-body homeostasis. After HFD feeding, AAV-CLSTN3 mice displayed an increased glucose intolerance as compared to the control mice in GTT, although insulin tolerance changed minimally in ITT (Figure 4F,G). Meanwhile, we observed that fasting insulin concentration was comparable between two groups (Figure 4H). This moderate change in GTT and no substantial change in overall ITT response may be explained by AAV-mediated local overexpression of CLSTN3 in iWAT. Highly inflammatory and fibrotic adipose tissue is usually associated with adverse changes in liver [37]. Therefore, we examined the liver of HFD-fed AAV-CLSTN3 mice for histological change, and noticed a pronounced fatty liver phenotype (Figure 4I); liver chemistry showed elevated triglyceride level (Figure 4J). Furthermore, serum concentrations of total cholesterol and triglyceride were higher in AAV-CLSTN3 mice (Figure 4K). Therefore, these data indicate that iWAT-specific CLSTN3 overexpression is associated with local, dysfunctional WAT expansion and liver steatosis, which in turn leads to moderate impairment of whole-body metabolism.
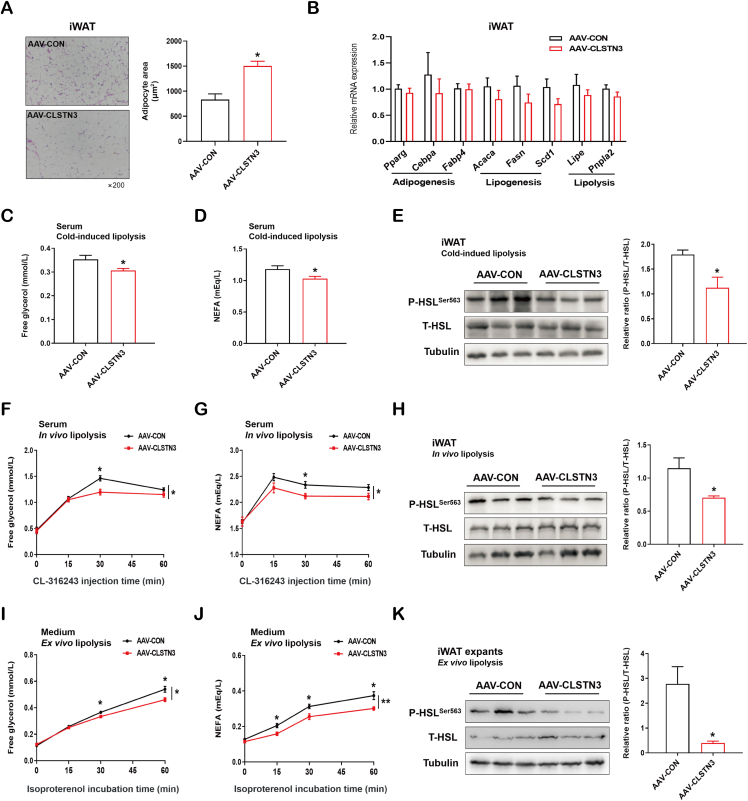
3.5. CLSTN3 attenuates catecholamine-stimulated lipolysis in vivo and ex vivo
The driving force for adipocyte hypertrophy after CLSTN3 induction is unclear. Surprisingly, we noted that CLSTN3-overexpressing iWAT displayed a rapid adipocyte enlargement after 4-week AAV administration (Figure 5A). This demonstrated that CLSTN3 overexpression is sufficient to drive adipocyte hypertrophy, even in the absence of HFD. Mechanistically, adipocyte size is regulated by the balance between lipogenesis and lipolysis [38]. Initially, we did not find any differences in the expression of genes that are critical for adipogenesis (Pparg, Cebpa, and Fabp4) in iWAT upon CLSTN3 overexpression, while there was a nonsignificant reduction in the expression of genes involved in lipogenesis (Acaca, Fasn, and Scd1) and lipolysis (Lipe and Pnpla2) (Figure 5B). Incidentally, HSL can be activated via its phosphorylation, and the level of P-HSL is used to assess the level of lipolysis [39]. Hence, we further explored the effect of CLSTN3 overexpression on HSL phosphorylation.
Figure 5.
CLSTN3 overexpression suppresses stimulated lipolysis in vivo and ex vivo. (A) Representative H&E staining images of iWAT sections of AAV-CON and AAV-CLSTN3 mice after 4-week AAV injection, and its quantification of adipocyte area. Scale bar, 50 μm. (B) Quantitative PCR analysis of mRNA expression of adipogenesis (Pparg, Cebpa, and Fabp4), lipogenesis (Acaca, Fasn, and Scd1) and lipolysis (Lipe and Pnpla2) related genes in iWAT from both groups (n = 5). (C and D) Free glycerol (C) and non-esterified fatty acid (NEFA) (D) levels in serum of AAV-CON and AAV-CLSTN3 mice upon 4-hour cold stress (n = 10). (E) Representative immunoblot analysis for the expression of phosphorylated HSL Ser563 (P-HSLSer563) and total HSL (T-HSL) in iWAT from both groups upon cold stress, and the quantification for the ratio of P-HSLSer563 to T-HSL expression. (F and G) Free glycerol (F) and NEFA (G) levels at different time points in serum of AAV-CON and AAV-CLSTN3 mice after β3-adrenoceptor agonist CL-316,243 injection (n = 7–8). (H) Immunoblot analysis and its quantification for the ratio of P-HSLSer563 to T-HSL expression in iWAT from both groups from in vivo lipolysis assay. (I and J) Free glycerol (I) and NEFA (J) levels at different time points in medium released from iWAT explants isolated from AAV-CON and AAV-CLSTN3 mice upon 10 μmol/L isoproterenol stimulation (n = 10). (K) Immunoblot analysis and its quantification for the ratio of P-HSLSer563 to T-HSL expression in iWAT from both groups from ex vivo lipolysis assay. Data are shown as mean ± SEM of biologically independent samples, and ∗p < 0.05, ∗∗p < 0.01.
Physiologically, lipolysis is coordinately controlled by the fasting/feed cycle and cold stress [40]. Firstly, we observed that CLSTN3 overexpression did not alter the levels of blood glucose and serum insulin either in fasted or fed state (Figure S4A and S4B). Also, it had no effect on fasting-induced lipolysis (Figure S4C and S4D). Specifically, in response to cold stress, the levels of free glycerol and NEFA in serum of CLSTN3-overexpressing mice were lower than that in the control mice (Figure 5C,D). We observed that P-HSL level in iWAT of CLSTN3-overexpressing mice was lower (Figure 5E). Moreover, we tested in vivo lipolytic capacity. We noticed that AAV-CON mice exhibited enhanced serum glycerol and NEFA levels at different time points in response to CL-316,243, while AAV-CLSTN3 mice failed to trigger marked glycerol and NEFA release at 30-min time point (Figure 5F,G). At the protein level, CLSTN3 overexpression attenuated the phosphorylation of HSL in iWAT (Figure 5H). Furthermore, our ex vivo lipolysis assay confirmed that iWAT explants obtained from AAV-CLSTN3 mice displayed an impaired response to isoproterenol-induced glycerol release from 30-min time point and NEFA release from 15-min time point (Figure 5I,J). Likewise, CLSTN3 overexpression reduced the ratio of P-HSL/T-HSL protein expression in iWAT explants (Figure 5K). Therefore, we conclude that CLSTN3 overexpression in iWAT causes adipocyte hypertrophy, at least in part, due to impaired catecholamine-stimulated lipolysis.
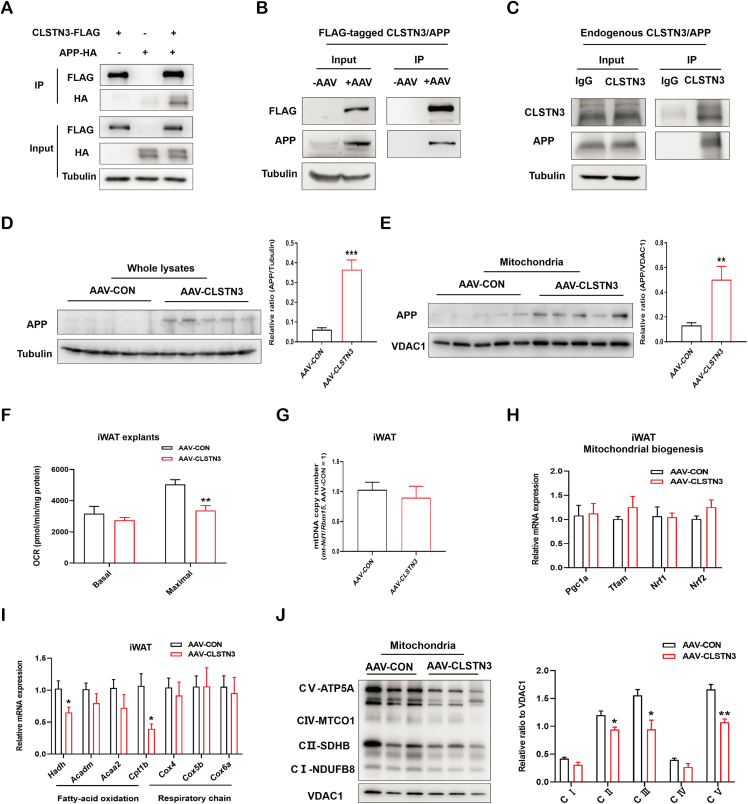
3.6. CLSTN3 modulates adipose mitochondrial function via its interaction with APP
Our observations raised a critical question regarding the mechanism by which CLSTN3 controls lipolysis in iWAT. Previous studies have established that CLSTN3 can interact with APP to form a complex that stabilizes APP metabolism in brain [21,22]. Initially, we examined the interaction between CLSTN3 and APP using immunoprecipitation assays. The interaction between overexpressed CLSTN3-FLAG and APP-HA was observed in 293T cells (Figure 6A). Endogenous APP could be immunoprecipitated with FLAG-tagged CLSTN3 in iWAT of AAV-CLSTN3 mice (Figure 6B). Moreover, endogenous CLSTN3 and APP exhibited a tight interaction in human SAT (Figure 6C). To examine whether the interaction affects APP level, we assessed its level in whole lysate of mouse iWAT. Notably, we observed that APP level was greatly upregulated in iWAT of AAV-CLSTN3 mice (Figure 6D). Obese conditions induce APP abnormal expression in WAT of mice, and excess APP is mis-targeted into mitochondria [41]. Consistent with this, we observed that APP level was substantially enriched in mitochondria isolated from CLSTN3-overexpressed iWAT (Figure 6E).
Figure 6.
CLSTN3 associates with APP and represses adipose mitochondrial function. (A) Immunoprecipitation of CLSTN3 with APP in 293T cells co-transfected with CLSTN3-FLAG and APP-HA plasmids for 48 h. Cell lysate (input) and pull-down (IP) of FLAG-tagged CLSTN3 were immunoblotted with indicated antibodies. (B) Immunoprecipitation of FLAG-tagged CLSTN3 and APP in iWAT from AAV-CON and AAV-CLSTN3 mice. (C) Immunoprecipitation of endogenous CLSTN3 and APP in human SAT. (D and E) Immunoblot analysis and the quantification for APP levels in whole-tissue lysate (D) and mitochondria (E) isolated from iWAT in AAV-CON and AAV-CLSTN3 mice; VDAC1, mitochondrial protein marker. (F) Ex vivo mitochondrial respiration in iWAT explants from AAV-CON and AAV-CLSTN3 mice; basal and maximal oxygen consumption rate (OCR) normalized to the protein content were presented (n = 5). (G) Mitochondrial DNA (mtDNA) copy number in iWAT from two groups (n = 5). (H) Quantitative PCR analysis of genes involved in mitochondrial biogenesis (Pgc1a, Tfam, Nrf1, and Nrf2) in iWAT from AAV-CON and AAV-CLSTN3 mice (n = 5). (I) Quantitative expression of genes involved in fatty-acid oxidation (Hadh, Acadm, Acaa2, and Cpt1b) and respiratory chain (Cox4, Cox5b, and Cox6a) in iWAT from AAV-CON and AAV-CLSTN3 mice (n = 5). (J) Immunoblot analysis and the quantification for crucial OXPHOS components, including NDUFB8 (CⅠ), SDHB (CII), UQCRC2 (CIII), MTCO1 (CⅣ), and ATP5A (CV), in mitochondria isolated from iWAT in AAV-CON and AAV-CLSTN3 mice. Data are shown as mean ± SEM of biologically independent samples, and ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Mitochondrial mis-localization of APP in WAT disrupts mitochondrial function and impairs lipolysis in mice [41]. This promotes us to examine whether the adverse effect of CLSTN3 on lipolysis is mediated through mitochondrial dysfunction caused by APP accumulation. Interestingly, we observed a substantial decline in maximal OCR of iWAT explants of AAV-CLSTN3 mice (Figure 6F). We additionally observed that CLSTN3 overexpression did not alter mtDNA copy number (Figure 6G) or gene expression levels of mitochondrial biogenesis regulators (Pgc1a, Tfam, Nrf1, and Nrf2) (Figure 6H). Furthermore, upon CLSTN3 overexpression, the majority of genes associated with fatty acid oxidation and respiration chain reaction remained unaffected, while Hadh and Cpt1b were downregulated (Figure 6I). Finally, we evaluated the protein levels of crucial OXPHOS components in mitochondria isolated from CLSTN3-overexpressed iWAT. Among these, we found that SDHB (CII), UQCRC2 (CIII), and ATP5A (CV) were significantly suppressed (Figure 6J). Hence, these observations indicate that CLSTN3 may cause mitochondrial dysfunction by interacting with APP and leading to APP enrichment in mitochondria.
4. Discussion
In the present study, we have established that CLSTN3 has a vital role in regulating lipolysis in adipose tissue, apart from its fundamental action in CNS. We combined human genomic dataset and eQTL analysis to provide the evidence that high CLSTN3 expression in adipose tissue correlates with the risk of human obesity. Our in vivo and ex vivo studies demonstrate that CLSTN3 enhancement causes iWAT dysfunction, partly due to impaired catecholamine-stimulated lipolysis.
Similar to other CLSTNs (CLSTN1 and CLSTN2), CLSTN3 is a type I transmembrane protein of cadherin superfamily. Although three members have similar molecular structure, CLSTN3 differs in function from the other two, because this protein has a unique C-terminus and shows a more prominent surface localization [16]. It promotes synapse development in CNS by interacting with α-neurexin [42,43]. Kim et al. showed novel physiological function of neuronal CLSTN3 in regulating energy homeostasis and bone metabolism in mice [44]. However, the relevance of CLSTN3 in peripheral tissues has not yet been studied thoroughly. Zeng et al. revealed a novel gene Clstn3b in mice, which shares the last two exons of Clstn3 gene and regulates systemic energy expenditure by controlling innervation of thermogenic adipose tissue. Clstn3b mRNA expression is specifically restricted to adipose tissue: it is most highly expressed in BAT, followed by iWAT and eWAT [33]. The information of Clstn3 expression in adipose tissue is lack. Our data showed that Clstn3 expression in mouse iWAT and eWAT is extremely low, while CLSTN3 was routinely expressed in human adipose tissue. Moreover, due to the shared sequence homology between Clstn3b and Clstn3 genes, their total transcripts have been identified in transcriptomic datasets of mouse and human fat [26,45]. Furthermore, the adipose-specificity of both Clstn3 and Clstn3b transcripts has been assessed separately in adipose depots of mice and humans under environmental cues [46]. In the current study, we observed that the expression pattern of CLSTN3 gene in human adipose tissue is different from that in mice. CLSTN3 transcript is routinely expressed in human adipose tissue and predominantly enriched in mature adipocyte fraction. Therefore, the overall function of CLSTN3 in adipose biology is worth exploring in depth.
Obesity is heritable, and it predisposes individuals to many metabolic diseases. GWAS have been performed till date to understand the genetic basis of the biological processes underlying obesity. It is worth noting that genetic loci associated with BMI overlap with genes involved in neurodevelopment, indicating a role of CNS, particularly the hypothalamus, in the regulation of body mass [47]. Adjusted-BMI loci are enriched for genes expressed in adipose depots and putative regulatory elements in adipocytes, and eQTL analysis provides insight to the potential pathophysiological mechanisms [48]. The non-coding variants may influence gene expression via chromatin modification, DNA accessibility and transcription factor binding in specific cell types and tissues [35]. For example, RIPK1 gene variants have been demonstrated to associate with human obesity; SNP rs5873855 at the RIPK1 intronic region disrupts one binding site for the transcriptional repressor E4BP4, and increases RIPK1 promoter activity and gene expression in adipose tissue [49]. SNP rs7296261 is located in the intron of CLSTN3 gene, and the variant may govern its transcriptional expression. However, the underlying gene regulatory elements and transcription factors that SNP rs7296261 influences need to be further defined. Here, we have demonstrated that rs7296261 is associated with high CLSTN3 expression in human adipose tissue and obesity risk. This implies that participants who are genetically susceptible to an increased expression of CLSTN3 tend to have a high risk of obesity. The data obtained from the examination of HFD-induced obese mice further support the role of CLSTN3 in adipose biology, wherein CLSTN3 overexpression results in a deterioration of WAT function and the onset of liver steatosis.
Body fat mass is determined by the balance between lipid storage and mobilization in adipocytes [50]. The physiological regulation of the release of fatty acid from triglyceride is stimulated by fasting or cold stress, and it occurs via the release of catecholamines from the sympathetic nerves [50,51]. Impaired lipolytic capacity commonly results in improved metabolic function, as reduced FFA liberation from adipose depots is thought to alleviate lipotoxicity in peripheral tissues, including liver [52,53]. An et al. revealed that mitochondrial dicarboxylate carrier mDIC prevents hepatic lipotoxicity by inhibiting white adipocyte lipolysis [53]. Meanwhile, they revealed that mitochondrial APP enhancement impairs catecholamine-induced lipolysis, thereby resulting in rapid adipocyte hypertrophy and liver steatosis [41]. Therefore, simple assessment on lipolysis in WAT may not be the determinant of ectopic fat deposition and metabolic dysfunction; however, which tissue becomes dysfunctional shortly should be considered. The early stage of inefficient subcutaneous adipocyte lipolysis predicts further weight gain and glucose intolerance in women [54]. Our data demonstrate that CLSTN3 overexpression leads to adipocyte hypertrophy and dysfunction rapidly due to mitochondrial dysfunction and lower catecholamine-stimulated lipolysis in iWAT depots, which in turn leads to the following development of liver steatosis and whole-body deficiency. In addition, the alteration of some adipokines directly contributes to the progression of metabolic liver diseases. For example, adiponectin is secreted exclusively from adipose tissue, and has been shown to reduce hepatic lipogenesis and increases β-oxidation to promote systemic energy homeostasis [55]. In the present study, CLSTN3-driven adipokines participating in adipocyte-liver crosstalk remain unknown yet.
It is well-known that mitochondrial dynamics regulate lipid storage and utilization [56]. However, the mediators between mitochondrial dysfunction after CLSTN3 overexpression and decreased catecholamine-stimulated lipolysis are currently unclear. There are certain modulators for the crosslink between mitochondrial function and lipolysis. LINC00473 is shuttled to the mitochondrial-lipid droplet interphase, and modulates mitochondrial responsiveness and lipolysis under catecholamine activation [57]. Beclin1 is the core molecule for macroautophagy machinery in adipose tissue, and it has critical roles in the maintenance of mitochondrial homeostasis and lipolysis in relation to β-adrenergic stimulation [58]. Therefore, the underlying inter-organelle communications after CLSTN3 overexpression are worthy to be further explored. Therapeutic silencing of CLSTN3 should be performed in adipose tissue to reinforce its function, while the expression level of Clstn3 is extremely low in mouse WAT, and interventions on human adipose tissue are challenging.
APP can be cleaved by proteases in amyloidogenic and non-amyloidogenic ways to produce a variety of short peptides, among which the role of amyloid β peptides in Alzheimer's disease has been intensively investigated [59]. Abnormal expression of full-length APP in peripheral tissues is associated with metabolic diseases [19]. Mitochondrial mis-localization of APP in adipocytes disrupts mitochondrial function, inhibits lipolysis, and promotes the occurrence of obesity in mice [41]. APP knockdown in adipocytes enhances mitochondrial respiration [60]. In our study, the similarities that were observed in the expression and function of CLSTN3 and APP in WAT suggest that they may share common signaling pathways in the regulation of adipose biology and systemic homeostasis. Therefore, we propose that the interaction between CLSTN3 and APP forms one of vital mechanisms underlying the pathological role of CLSTN3 in controlling adipocyte mitochondrial function.
There are several limitations of our study. First, the effect of CLSTN3 on WAT dysfunction are proved through AAV-mediated overexpression, and more should focus on the comprehensive role of CLSTN3 in adipocyte-specific CLSTN3 transgenic mice. Second, there is a correlation between CLSTN3 expression and APP localization to mitochondria, while the role of CLSTN3 in modulating WAT function through APP translocation pathway needs to be thoroughly investigated. Last, the associations of CLSTN3 variant rs7296261 on metabolic traits and its expression in adipose tissue need to be strengthened in a human cohort with the lean and varying degree of obesity in future studies.
In conclusion, our work suggests that the presence of CLSTN3 gene variant is correlated with high CLSTN3 expression in human adipose tissue, which in turn is associated with unfavorable phenotypes. In the context of obesity, we have demonstrated a novel role of CLSTN3 in adipose mitochondrial function, catecholamine-induced lipolysis, and adipocyte hypertrophy. We believe that CLSTN3 may be a therapeutic target for the treatment of obesity and associated metabolic diseases.
Funding
This study was financially supported by grants from National Key Research and Development Program of China (2019YFA0904501) and National Natural Science Foundation of China (No. 81974122).
Author contributions
YY, CH, and JH conceived and designed the study. NB, XL, and LJ performed the experiments, collected and analyzed the results. MA, JM, FH, YX, JS, JX, and RZ assisted with experiments and data analysis. NB and YY wrote the paper. CH and JH reviewed the manuscript. All authors approved the final version of the manuscript to be published.
Acknowledgements
We thank Professor Jiahuai Han at Xiamen University for providing human CLSTN3 plasmid.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101531.
Contributor Information
Junfeng Han, Email: tjhjf@163.com.
Cheng Hu, Email: alfredhc@sjtu.edu.cn.
Ying Yang, Email: yangyingsh@sjtu.edu.cn.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Kusminski C.M., Bickel P.E., Scherer P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nature Reviews Drug Discovery. 2016;15(9):639–660. doi: 10.1038/nrd.2016.75. [DOI] [PubMed] [Google Scholar]
- 2.Tchoukalova Y.D., Votruba S.B., Tchkonia T., Giorgadze N., Kirkland J.L., Jensen M.D. Regional differences in cellular mechanisms of adipose tissue gain with overfeeding. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(42):18226–18231. doi: 10.1073/pnas.1005259107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tandon P., Wafer R., Minchin J.E.N. Adipose morphology and metabolic disease. Journal of Experimental Biology. 2018;221(Pt Suppl 1):jeb164970. doi: 10.1242/jeb.164970. [DOI] [PubMed] [Google Scholar]
- 4.Crewe C., An Y.A., Scherer P.E. The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis. Journal of Clinical Investigation. 2017;127(1):74–82. doi: 10.1172/JCI88883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lönn M., Mehlig K., Bengtsson C., Lissner L. Adipocyte size predicts incidence of type 2 diabetes in women. The FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 2010;24(1):326–331. doi: 10.1096/fj.09-133058. [DOI] [PubMed] [Google Scholar]
- 6.Stunkard A.J., Foch T.T., Hrubec Z. A twin study of human obesity. JAMA. 1986;256(1):51–54. [PubMed] [Google Scholar]
- 7.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Y., Loos R.J. Obesity genomics: assessing the transferability of susceptibility loci across diverse populations. Genome Medicine. 2013;5(6):55. doi: 10.1186/gm459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun J.S., Ko S.H. Current trends in epidemiology of cardiovascular disease and cardiovascular risk management in type 2 diabetes. Metabolism Clinical and Experimental. 2021;123 doi: 10.1016/j.metabol.2021.154838. [DOI] [PubMed] [Google Scholar]
- 10.Loos R.J., Yeo G.S. The bigger picture of FTO: the first GWAS-identified obesity gene. Nature Reviews Endocrinology. 2014;10(1):51–61. doi: 10.1038/nrendo.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science (New York, N.Y.) 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khera A.V., Chaffin M., Wade K.H., Zahid S., Brancale J., Xia R., et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019;177(3):587–596. doi: 10.1016/j.cell.2019.03.028. e589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T., Jia W., Hu C. Advancement in genetic variants conferring obesity susceptibility from genome-wide association studies. Frontiers of Medicine. 2015;9(2):146–161. doi: 10.1007/s11684-014-0373-8. [DOI] [PubMed] [Google Scholar]
- 14.Landgraf K., Klöting N., Gericke M., Maixner N., Guiu-Jurado E., Scholz M., et al. The obesity-susceptibility gene TMEM18 promotes adipogenesis through activation of PPARG. Cell Reports. 2020;33(3) doi: 10.1016/j.celrep.2020.108295. [DOI] [PubMed] [Google Scholar]
- 15.Fathzadeh M., Li J., Rao A., Cook N., Chennamsetty I., Seldin M., et al. FAM13A affects body fat distribution and adipocyte function. Nature Communications. 2020;11(1):1465. doi: 10.1038/s41467-020-15291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hintsch G., Zurlinden A., Meskenaite V., Steuble M., Fink-Widmer K., Kinter J., et al. The calsyntenins--a family of postsynaptic membrane proteins with distinct neuronal expression patterns. Molecular and cellular neurosciences. 2002;21(3):393–409. doi: 10.1006/mcne.2002.1181. [DOI] [PubMed] [Google Scholar]
- 17.de Ramon Francàs G., Alther T., Stoeckli E.T. Calsyntenins are expressed in a dynamic and partially overlapping manner during neural development. Frontiers in Neuroanatomy. 2017;11:76. doi: 10.3389/fnana.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Z., Wang Y., Chen F., Tong H., Reddy M.V., Luo L., et al. Calsyntenin-3 molecular architecture and interaction with neurexin 1alpha. Journal of Biological Chemistry. 2014;289(50):34530–34542. doi: 10.1074/jbc.M114.606806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y., Wang Q., Chen S., Xu C. Functions of amyloid precursor protein in metabolic diseases. Metabolism Clinical and Experimental. 2021;115 doi: 10.1016/j.metabol.2020.154454. [DOI] [PubMed] [Google Scholar]
- 20.Zheng H., Koo E.H. Biology and pathophysiology of the amyloid precursor protein. Molecular Neurodegeneration. 2011;6(1):27. doi: 10.1186/1750-1326-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araki Y., Tomita S., Yamaguchi H., Miyagi N., Sumioka A., Kirino Y., et al. Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid beta-protein precursor metabolism. Journal of Biological Chemistry. 2003;278(49):49448–49458. doi: 10.1074/jbc.M306024200. [DOI] [PubMed] [Google Scholar]
- 22.Dinamarca M.C., Raveh A., Schneider A., Fritzius T., Fruh S., Rem P.D., et al. Complex formation of APP with GABAB receptors links axonal trafficking to amyloidogenic processing. Nature Communications. 2019;10(1):1331. doi: 10.1038/s41467-019-09164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao Y., Ma X., Yang R., Wang F., Hao Y., Dou J., et al. Inverse relationship between serum osteocalcin levels and visceral fat area in Chinese men. Journal of Clinical Endocrinology & Metabolism. 2013;98(1):345–351. doi: 10.1210/jc.2012-2906. [DOI] [PubMed] [Google Scholar]
- 24.He Z., Zhang R., Jiang F., Zhang H., Zhao A., Xu B., et al. FADS1-FADS2 genetic polymorphisms are associated with fatty acid metabolism through changes in DNA methylation and gene expression. Clinical Epigenetics. 2018;10(1):113. doi: 10.1186/s13148-018-0545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Jin L., Jiang F., Yan J., Lu Y., Yang Q., et al. Mutations of NRG4 contribute to the pathogenesis of non-alcoholic fatty liver disease and related metabolic disorders. Diabetes. 2021;70(10):2213–2224. doi: 10.2337/db21-0064. [DOI] [PubMed] [Google Scholar]
- 26.Jespersen N.Z., Feizi A., Andersen E.S., Heywood S., Hattel H.B., Daugaard S., et al. Heterogeneity in the perirenal region of humans suggests presence of dormant brown adipose tissue that contains brown fat precursor cells. Molecular Metabolism. 2019;24:30–43. doi: 10.1016/j.molmet.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y.T., Li L.Z., Yang Y.L., Yin X., Liu Q., Zhang L., et al. Succinate induces aberrant mitochondrial fission in cardiomyocytes through GPR91 signaling. Cell Death & Disease. 2018;9(6):672. doi: 10.1038/s41419-018-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu B., Liu C., Zhang H., Zhang R., Tang M., Huang Y., et al. Skeletal muscle-targeted delivery of Fgf6 protects mice from diet-induced obesity and insulin resistance. JCI insight. 2021;6(19) doi: 10.1172/jci.insight.149969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alimujiang M., Sun J., Chen S., Bai N., Chen S., Hu F., et al. Survivin is essential for thermogenic program and metabolic homeostasis in mice. Molecular Metabolism. 2022;58 doi: 10.1016/j.molmet.2022.101446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ju L., Zhang X., Deng Y., Han J., Yang J., Chen S., et al. Enhanced expression of Survivin has distinct roles in adipocyte homeostasis. Cell Death & Disease. 2017;8(1) doi: 10.1038/cddis.2016.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunham-Snary K.J., Sandel M.W., Westbrook D.G., Ballinger S.W. A method for assessing mitochondrial bioenergetics in whole white adipose tissues. Redox Biology. 2014;2:656–660. doi: 10.1016/j.redox.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai N., Ma J., Alimujiang M., Xu J., Hu F., Xu Y., et al. Bola3 regulates beige adipocyte thermogenesis via maintaining mitochondrial homeostasis and lipolysis. Frontiers in Endocrinology. 2020;11 doi: 10.3389/fendo.2020.592154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng X., Ye M., Resch J.M., Jedrychowski M.P., Hu B., Lowell B.B., et al. Innervation of thermogenic adipose tissue via a calsyntenin 3beta-S100b axis. Nature. 2019;569(7755):229–235. doi: 10.1038/s41586-019-1156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magkos F., Lee M.H., Lim M., Cook A.R., Chhay V., Loh T.P., et al. Dynamic assessment of insulin secretion and insulin resistance in Asians with prediabetes. Metabolism Clinical and Experimental. 2021;128 doi: 10.1016/j.metabol.2021.154957. [DOI] [PubMed] [Google Scholar]
- 35.Wong, A.K., Sealfon, R.S.G., Theesfeld, C.L., Troyanskaya, O.G., 2021. Decoding disease: from genomes to networks to phenotypes. Nature Reviews Genetics 22(12):774-790. [DOI] [PubMed]
- 36.Consortium T.G. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (New York, N.Y.) 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byrne C.D., Targher G. NAFLD: a multisystem disease. Journal of Hepatology. 2015;62(1 Suppl):S47–S64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Haczeyni F., Bell-Anderson K.S., Farrell G.C. Causes and mechanisms of adipocyte enlargement and adipose expansion. Obesity Reviews : An Official Journal of the International Association for the Study of Obesity. 2018;19(3):406–420. doi: 10.1111/obr.12646. [DOI] [PubMed] [Google Scholar]
- 39.Strålfors P., Belfrage P. Phosphorylation of hormone-sensitive lipase by cyclic AMP-dependent protein kinase. Journal of Biological Chemistry. 1983;258(24):15146–15152. [PubMed] [Google Scholar]
- 40.Grabner G.F., Xie H., Schweiger M., Zechner R. Lipolysis: cellular mechanisms for lipid mobilization from fat stores. Nature Metabolism. 2021;3(11):1445–1465. doi: 10.1038/s42255-021-00493-6. [DOI] [PubMed] [Google Scholar]
- 41.An Y.A., Crewe C., Asterholm I.W., Sun K., Chen S., Zhang F., et al. Dysregulation of amyloid precursor protein impairs adipose tissue mitochondrial function and promotes obesity. Nature Metabolism. 2019;1(12):1243–1257. doi: 10.1038/s42255-019-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettem K.L., Yokomaku D., Luo L., Linhoff M.W., Prasad T., Connor S.A., et al. The specific alpha-neurexin interactor calsyntenin-3 promotes excitatory and inhibitory synapse development. Neuron. 2013;80(1):113–128. doi: 10.1016/j.neuron.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Um J.W., Pramanik G., Ko J.S., Song M.Y., Lee D., Kim H., et al. Calsyntenins function as synaptogenic adhesion molecules in concert with neurexins. Cell Reports. 2014;6(6):1096–1109. doi: 10.1016/j.celrep.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S.J., Jeong Y.T., Jeong S.R., Park M., Go H.S., Kim M.Y., et al. Neural regulation of energy and bone homeostasis by the synaptic adhesion molecule Calsyntenin-3. Experimental & Molecular Medicine. 2020;52(5):793–803. doi: 10.1038/s12276-020-0419-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S.Q., Niu Q., Ju L.P., Alimujiang M., Yan H., Bai N.N., et al. Predicted secreted protein analysis reveals synaptogenic function of Clstn3 during WAT browning and BAT activation in mice. Acta Pharmacologica Sinica. 2019;40(8):999–1009. doi: 10.1038/s41401-019-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plucinska K., Jespersen N.Z., Brown E.L., Petersen P.S., Rupar K., Nielsen S., et al. Calsyntenin 3beta is dynamically regulated by temperature in murine Brown adipose and marks human multilocular fat. Frontiers in Endocrinology. 2020;11 doi: 10.3389/fendo.2020.579785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karunakaran D., Turner A.W., Duchez A.C., Soubeyrand S., Rasheed A., Smyth D., et al. RIPK1 gene variants associate with obesity in humans and can be therapeutically silenced to reduce obesity in mice. Nature Metabolism. 2020;2:1113–1125. doi: 10.1038/s42255-020-00279-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arner P. Human fat cell lipolysis: biochemistry, regulation and clinical role. Best practice & research. Clinical endocrinology & metabolism. 2005;19(4):471–482. doi: 10.1016/j.beem.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Chouchani E.T., Kajimura S. Metabolic adaptation and maladaptation in adipose tissue. Nature Metabolism. 2019;1(2):189–200. doi: 10.1038/s42255-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samocha-Bonet D., Chisholm D.J., Tonks K., Campbell L.V., Greenfield J.R. Insulin-sensitive obesity in humans - a 'favorable fat' phenotype? Trends in Endocrinology and Metabolism: Trends in Endocrinology and Metabolism. 2012;23(3):116–124. doi: 10.1016/j.tem.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 53.An Y.A., Chen S., Deng Y., Wang Z.V., Funcke J.B., Shah M., et al. The mitochondrial dicarboxylate carrier prevents hepatic lipotoxicity by inhibiting white adipocyte lipolysis. Journal of Hepatology. 2021;75(2):387–399. doi: 10.1016/j.jhep.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arner P., Andersson D.P., Backdahl J., Dahlman I., Ryden M. Weight gain and impaired glucose metabolism in women are predicted by inefficient subcutaneous fat cell lipolysis. Cell Metabolism. 2018;28(1):45–54 e43. doi: 10.1016/j.cmet.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Stern J.H., Rutkowski J.M., Scherer P.E. Adiponectin, leptin, and fatty acids in the maintenance of metabolic homeostasis through adipose tissue crosstalk. Cell Metabolism. 2016;23(5):770–784. doi: 10.1016/j.cmet.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benador I.Y., Veliova M., Liesa M., Shirihai O.S. Mitochondria bound to lipid droplets: where mitochondrial dynamics regulate lipid storage and utilization. Cell Metabolism. 2019;29(4):827–835. doi: 10.1016/j.cmet.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran K.V., Brown E.L., DeSouza T., Jespersen N.Z., Nandrup-Bus C., Yang Q., et al. Human thermogenic adipocyte regulation by the long noncoding RNA LINC00473. Nature Metabolism. 2020;2(5):397–412. doi: 10.1038/s42255-020-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Son Y., Cho Y.K., Saha A., Kwon H.J., Park J.H., Kim M., et al. Adipocyte-specific Beclin1 deletion impairs lipolysis and mitochondrial integrity in adipose tissue. Molecular Metabolism. 2020;39 doi: 10.1016/j.molmet.2020.101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wurtman R. Biomarkers in the diagnosis and management of Alzheimer's disease. Metabolism Clinical and Experimental. 2015;64(3 Suppl 1):S47–S50. doi: 10.1016/j.metabol.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 60.Bjune J.I., Haugen C., Gudbrandsen O., Nordbø O.P., Nielsen H.J., Våge V., et al. IRX5 regulates adipocyte amyloid precursor protein and mitochondrial respiration in obesity. International Journal of Obesity. 2019;43(11):2151–2162. doi: 10.1038/s41366-018-0275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.