Figure 1. Regulated cargo ubiquitylation to promotes autophagic clearance of damaged mitochondria and intracellular bacteria.
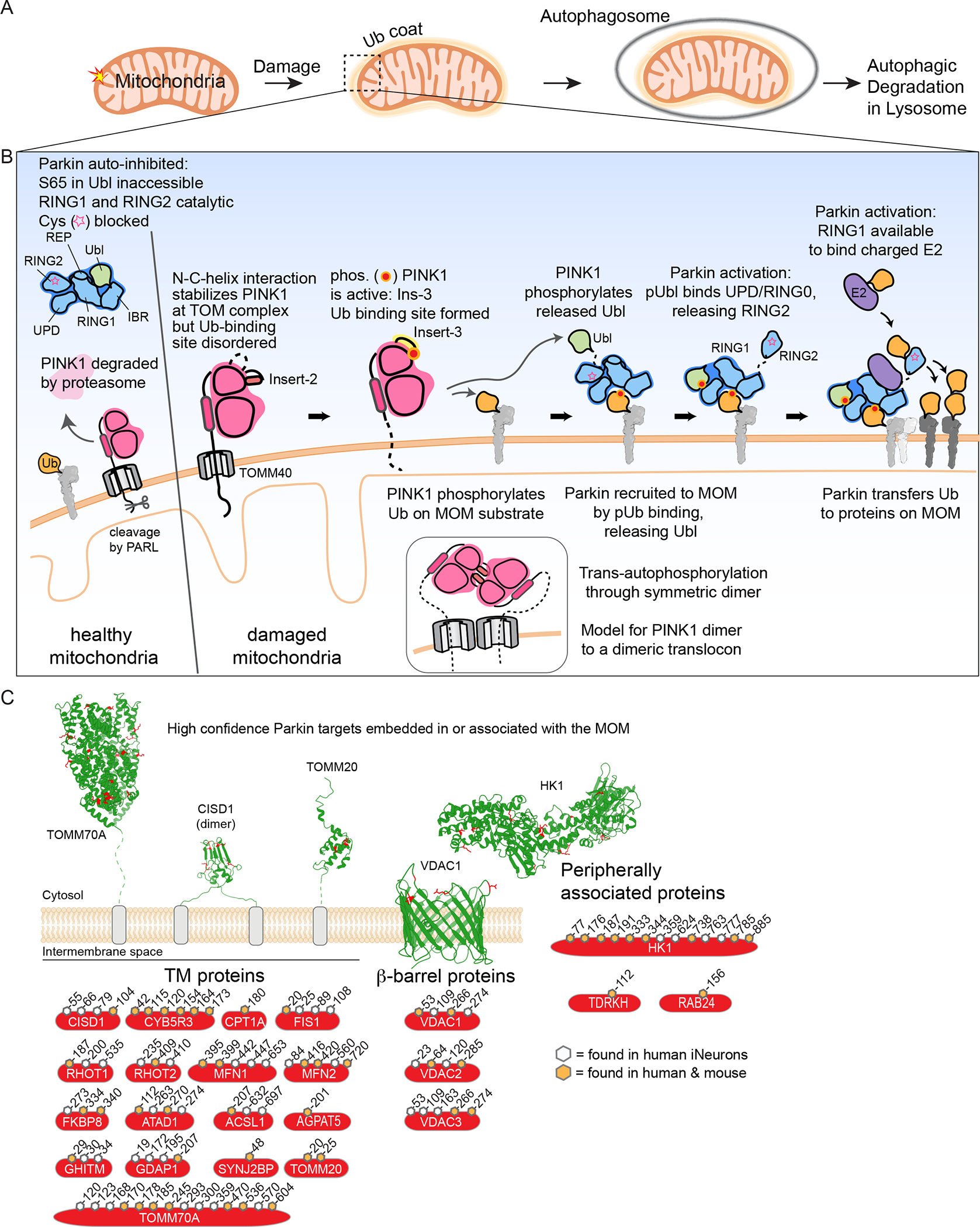
(A) Schematic of Ub-dependent mitophagy. Damaged mitochondria elicits a signal resulting in creation of a Ub “coat” surrounding the mitochondria. This Ub coat functions to promote formation of an autophagosome, which then can fuse with the lysosome to promote mitochondrial clearance.
(B) A feed-forward mechanism for Parkin and PINK1-dependent mitophagy. In cells with healthy mitochondria, cytosolic Parkin is in an auto-inhibited form and PINK1 is rapidly degraded. In response to mitochondrial damage, PINK1 is stabilized on the MOM in association with the translocon and trans-phosphorylates to generate a binding site for both Ub and the Parkin Ubl. MOM-tethered PINK1 can phosphorylate pre-existing Ub in proximity to the outer membrane, which can then bind Parkin to release its Ubl, making it available for phosphorylation by PINK1. Phosphorylated and activated Parkin can ubiquitylate numerous proteins on the MOM, thereby promoting downstream steps in mitophagy. Insets schematically depict dimeric PINK1 as an intermediate in trans-autophosphorylation, possibly templated by association with a dimeric translocon.
(C) High-confidence membrane-bound Parkin substrates, classified by membrane interaction type: single or double transmembrane segments, β-barrel proteins, and peripherally associated proteins. Examples displayed with identified ubiquitylated Lys residues shown in red and the cytoplasmic face of the MOM facing up. The high confidence ubiquitylation sites shown are based on biochemical studies in human and mouse cells in the context of endogenous Parkin in either human induced neurons (Ordureau et al., 2020) or primary mouse neurons (Antico et al., 2021). Sites indicated are high confidence sites in human proteins and orange-filled hexagons indicate sites that are also observed to be ubiquitylation by Parkin in mouse primary neurons. PDB files used: TOMM70A (2GW1), CISD1 (3EW0), TOMM20 (1OM2), VDAC1 (2JK4), and HK1 (1CZA).
