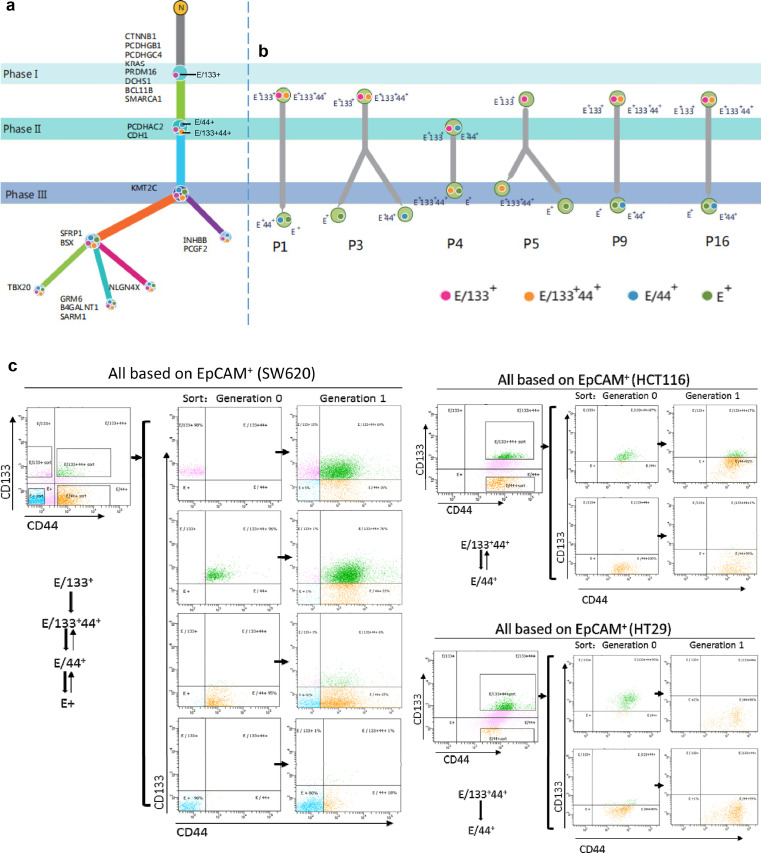
Figure 4.
Clonal evolution of CRCICs and CRCCs of scWES and TES data, and its validation of CRC cell lines. a. Clonal evolution of the scWES data. We used the OncoNEM method to analyze the clonal evolution of all single cells. The three phases are marked as I, II, III, showing the evolution of each step with specific gene mutations. E/133+ cells were marked as solid pink circles, while E/133+44+ cells were solid yellow circles, E/44+ cells were solid blue circles, and E+ cells were solid green circles. E/133+ cells are the original CRC cells. b. The targeted sequencing panel that included all mutant genes of scWES data were used to analyze the clonal evolution of other six patients. The cancer cells of these six patients were origin from cancer initiating cells (E/133+44+ or E/133+). The E/133+44+, E/44+, E/133+ and E+ cells of each patient were sorted by FACS. The three phases are marked as I, II, III, showing the evolution of each step with specific gene mutations. E/133+ cells are the original CRC cells. c. E/133+44+, E/44+, E/133+, and E+ cells from cell lines SW620, HCT116, and HT29 were sorted by FACS and then cultured. After one generation, E/133+ cells could grow into E/133+44+, E/44+, and E+ cells.