Abstract
Polygenic risk scores (PRS) represent an individual’s summed genetic risk for a trait and can serve as biomarkers for disease. Less is known about the utility of PRS as a means to quantify genetic risk for substance use disorders (SUDs) than for many other traits. Nonetheless, the growth of large, electronic health record-based biobanks makes it possible to evaluate the association of SUD PRS with other traits. We calculated PRS for smoking initiation, alcohol use disorder (AUD), and opioid use disorder (OUD) using summary statistics from the Million Veteran Program sample. We then tested the association of each PRS with its primary phenotype in the Penn Medicine BioBank (PMBB) using all available genotyped participants of African or European ancestry (AFR and EUR, respectively) (N=18,612). Finally, we conducted phenome-wide association analyses (PheWAS) separately by ancestry and sex to test for associations across disease categories. Tobacco use disorder was the most common SUD in the PMBB, followed by AUD and OUD, consistent with the population prevalence of these disorders. All PRS were associated with their primary phenotype in both ancestry groups. PheWAS results yielded cross-trait associations across multiple domains, including psychiatric disorders and medical conditions. SUD PRS were associated with their primary phenotypes, however they are not yet predictive enough to be useful diagnostically. The cross-trait associations of the SUD PRS are indicative of a broader genetic liability. Future work should extend findings to additional population groups and for other substances of abuse.
Keywords: electronic health record, genome-wide association study, phenome-wide association study, polygenic risk score, substance use disorders
Introduction
Substance use disorders (SUDs) are prevalent and costly to society. In 2019, 7.8% of U.S. adults had a current (past-year; PY) SUD.1 These disorders are associated with a host of negative outcomes, including poorer quality of life and increased mortality risk.2-4 The most common SUDs are tobacco use disorder (TUD; 26.9% PY) and alcohol use disorder (AUD; 13.9% PY).5,6 Although opioid use disorder (OUD) is less prevalent (0.6% PY) than either TUD or AUD, in 2019, 10.1 million individuals endorsed PY opioid misuse,1 which is of particular public health concern due to the associated high risk of fatal opioid overdose.7
Evidence from twin studies support a genetic component to the etiology of SUDs, and genome-wide association studies (GWAS) have identified dozens of variants contributing to SUD risk.8-10 Nonetheless, because SUDs are polygenic traits, no single variant accounts for more than a small portion of the variance in risk for developing an SUD. Polygenic risk scores (PRS) calculated from the weighted effect size of the GWAS-derived variants associated with a trait provide an aggregate measure of common genetic risk.11 PRS have been used to estimate the risk of medical diseases,12 health outcomes,13 and psychiatric disorders.14
Additionally, PRS have been used to investigate the genetic overlap between multiple phenotypes, which can help to evaluate overlapping and unique features of SUDs, that share symptomatology and, to some extent, underlying genetic architecture.15 Consistent with these features, a study of 11 different psychiatric and SUDs GWAS yielded strong genetic correlations between alcohol and tobacco dependence and other psychiatric disorders.16
Electronic health records (EHRs) are integral to the investigation of co-occurring disorders, as they provide a wealth of longitudinal phenotypic information that is often richer than that collected in clinical trials or cross-sectional studies. This longitudinal perspective, which provide repeated measures of a trait over long periods of time, can thereby increase confidence in the accuracy of diagnoses. Biobanks that provide genetic data linked with EHRs have been the basis for the development of phenome-wide association studies (PheWAS).17 PheWAS can use genetic data, such as a genetic variant or a PRS, to query phenotypic information (e.g., medical or psychiatric diagnoses, environmental factors) to identify associations, thus potentially illuminating shared genetic etiology or yet undiscovered phenotypic comorbidity. Although the use of GWAS and PheWAS has led to the identification of thousands of disease-associated variants, the vast majority of this literature comes from studies of individuals of European ancestry, underscoring the substantial presence of health disparities in genetic research. Women have also historically been underrepresented in medical research. The Penn Medicine BioBank (PMBB), like a number of other biobanks, is well suited to address these gaps in the genetics literature given the heterogeneity of its participants, who are drawn from a large urban center with a diverse population.
Using available summary statistics, we calculated PRS for three common substance-related traits; smoking initiation, AUD, and OUD, in both African-ancestry (AFR) and European-ancestry (EUR) individuals and examined their performance and their phenotypic associations in the PMBB. We also performed PheWAS to identify cross-trait associations and phenotypic overlap in genetic liability for SUDs, and a secondary PheWAS where the primary phenotype was covaried to determine whether identified associations persist when controlling for the index phenotype. Lastly, we conducted PheWAS separately in men and women to test whether the associations differed by sex.
Methods
Participants
Patients in the Penn Medicine BioBank (PMBB) were ascertained at the time of a medical appointment in the University of Pennsylvania Health System. At enrollment, participants gave informed consent and provided either a blood or tissue sample and permission to access their EHR information. As of July 2020, there were 63,177 individuals in PMBB, 21,263 of whom had genome-wide genotyping.
Genotyping and Quality Control
DNA was extracted from blood and samples were genotyped in four batches: 1) N=10,867 using the Illumina InfiniumOmniExpress-24v1-2_A1 (OMNI) at the Regeneron Genetics Center (RGC); 2) N=5,676, using the Global Screening Array (GSA) V1 at the Center for Applied Genomics (CAG) at the Children’s Hospital of Philadelphia; 3) N=2,972, using the GSA V2 chip at CAG; and 4) N=16,940 using the GSA V2 chip, at RGC. Some samples were genotyped multiple times. To impute unique samples, they were combined by genotyping chip into two batches (3 GSA chips into a GSA batch and 1 OMNI chip into an OMNI batch), with preference for unique samples in the following order: GSA V2, GSA V1, OMNI. Each batch underwent quality control separately prior to imputation. Samples were removed if the genotyping call rate was <90% or if genotyped sex did not match reported gender, leaving 20,079 unique GSA samples and 1,111 unique OMNI samples. SNPs were removed if the call rate was <95%, leaving N=622,717 SNPs in GSA and N=640,714 SNPs in OMNI.
Phasing and Imputation
Prior to imputation, SNPs were matched to the appropriate strand and those that were palindromic, not matched to the reference panel, with allele frequency difference >0.2, or with differing alleles were removed. Genotypes were phased (using EAGLE) and imputed to the TOPMed Reference Panel (Freeze 5) on the TOPMed Imputation server.18 In the GSA batch, 151,143,913 SNPs were imputed with R2>0.3. In the OMNI batch, 46,386,520 SNPs were imputed with R2>0.3.
Imputed genotypes underwent quality control (R2>0.3, marker call rate>95%, sample call rate>90%, MAF>0.5%) using PLINK v1.90. Related individuals were identified using a graph-based algorithm after applying a Pi-HAT threshold of 0.25, and the sample most closely related to multiple other samples was removed (n=738). Following removal of related individuals, principal component (PC) analysis was conducted using the Eigensoft smartpca module.19 We performed quantitative discriminant analysis to determine genetically informed ancestry, using 1000 Genomes, phase 320 as a training set and PMBB as a testing set.
Polygenic Risk Scores
We retained SNPs present in the HapMap reference panel (n=1,120,629) and merged the two batches into a single dataset. We extracted AFR (n=8,276) and EUR (n=10,473) individuals and used PLINK v1.90 to calculate ancestry-specific PCs to use as covariates. Summary statistics for smoking initiation (current vs. never smokers, Contrast I),10 AUD (AFR8; EUR21), and OUD9 were obtained from the Million Veteran Program (MVP) (Supplemental Table S1). We chose the MVP sample as our main source of GWAS summary statistics as it is the largest available source for these phenotypes in AFR individuals, and is one of the largest sources for EUR individuals. Ancestry specific PRS were calculated using PRS-CS,22 with AFR GWAS summary statistics and the 1000G AFR LD reference panel used in the PMBB AFR individuals, and the EUR GWAS summary statistics and the 1000G EUR LD reference panel used in the PMBB EUR individuals. Phi for all traits was fixed to 1e-2, with default thresholds used for everything else.
Phenotypes
International Classification of Diseases (ICD)-9 and ICD-10 data for 63,199 individuals in PMBB were extracted from the EHR and used to assign both primary phenotypes and phecodes. To increase the accuracy of diagnostic data, encounter type was filtered to include only records representing physician encounters, as previously described.14 This left 11,966,749 encounters (5,731,365 ICD-9; 6,235,384 ICD-10) for 63,177 individuals. Primary phenotypes were assigned based on the presence or absence of ICD codes. ICD codes for TUD, AUD, and OUD were selected based on clinician expertise (Supplemental Table S2). We classified primary phenotypes in two ways: a less stringent definition (requiring the presence of ≥1 or more ICD codes for the phenotype), and a stringent definition (requiring the presence of ≥1 ICD codes as inpatient, or ≥2 as outpatient). To create a dataset for PheWAS analysis in the PMBB, we aggregated ICD-9 codes to phecodes using the phecode ICD-9 map 1.2 and ICD-10 codes using the phecode ICD-10-CM map 1.2 (beta). Individuals were considered cases for the phecode if it was assigned on at least 2 unique dates, controls if they have no instance of the phecode, and ‘other/missing’ if they had one instance or a related phecode.23 In total, 1814 phecodes were generated. The final dataset included 18,612 individuals (8,235 AAs, 10,377 EAs) with complete genotype, phenotype, and covariate data.
Statistical Analysis
Individuals with a given SUD and those without that SUD were compared on demographics and comorbid medical conditions via chi-square for categorical data and t-tests for continuous data. PRS were standardized with mean=0 and standard deviation (SD)=1. Logistic regression was used to test for the association of each psychiatric risk score with the primary phenotype, which was considered significant at p<0.05, and to estimate the odds ratio (OR) for cases by comparing the top quintile of polygenic risk to the remaining quintiles of risk. We used TUD as the primary phenotype for smoking initiation, as it was the most similar one available in the PMBB EHR. The PheWAS analysis was performed in R using logistic regression models in which each PRS was the independent variable; phecodes were the dependent variables; and age, sex, and the first 10 PCs were covariates. We performed a second PheWAS that adjusted for the primary phenotype by including it as a covariate in the regression model and a third PheWAS that tested for sex differences. Phecodes with >100 cases (n=583 for AFR; n=477 for EUR) were tested (see Supplement 2). As a different number of Phecodes were tested in each ancestry, Bonferroni-corrected phenome-wide significance thresholds to account for multiple testing were p<8.59x10−5 for AFR and p<1.05x10−4 for EUR.
Results
Phenotypes in PMBB
Participants’ demographics and information on their comorbid conditions are presented in Table 1 for the genotyped sample (n=18,612) by SUD and ancestry. The sample was 53% male, with a mean age of 65 (SD=16.7) years, and a mean of 61 (SD=9.9) encounters in the EHR. As anticipated due to the initial PMBB recruitment strategy, there were high rates of comorbid circulatory system (82%), endocrine/metabolic (73.5%) and respiratory system (51%) diagnoses. Notably, individuals with SUDs had significantly higher rates of comorbid conditions, irrespective of SUD or ancestry.
Table 1.
Demographic information and prevalence of key PheCode comorbidities
| PMBB (18612) |
AFR | EUR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TUD + (2769) |
TUD − (5466) |
AUD + (592) |
AUD − (7643) |
OUD + (170) |
OUD − (8065) |
TUD + (3883) |
TUD − (6494) |
AUD + (424) |
AUD − (9953) |
OUD + (95) |
OUD − (10282) |
||
| Sex, n (% male) | 9834 (52.8) |
2355 (45.3) |
1822 (33.3)** |
371 (62.7) |
2706 (35.4)** |
76 (44.7) |
3001 (37.2)* |
2801 (72.1) |
3956 (60.9)** |
354 (83.5) |
6403 (64.3)** |
69 (72.6) |
6688 (65.0) |
| Age, M (SD) | 64.9 (16.7) |
61.83 (14.2) |
54.26 (16.8)** |
60.79 (12.3) |
56.5 (16.6)** |
58.04 (12.5) |
56.78 (16.5) |
73.4 (12.1) |
70.1 (14.7)** |
68.4 (11.5) |
71.5 (14.0)** |
62.2 (13.1) |
71.4 (13.9)** |
| # Encounters, M (SD) | 60.7 (69.9) |
96.7 (85.6) |
66.0 (65.3)** |
103.2 (86.1) |
74.2 (72.8)** |
140.4 (106.4) |
75.0 (72.8)** |
53.9 (68.2) |
44.9 (60.6)** |
72.8 (79.8) |
47.2 (62.7)** |
116.1 (113.9) |
47.7 (62.7)** |
| Circulatory System, n(%) | 15327 (82.4) |
2431 (87.8) |
3668 (67.1)** |
523 (88.3) |
5576 (73.0)** |
154 (90.6) |
5945 (73.7)** |
3701 (95.3) |
5527 (85.1)** |
387 (91.3) |
8841 (88.8) |
93 (97.9) |
9135 (88.8)* |
| Endocrine/Metabolic, n(%) | 13686 (73.5) |
2372 (85.7) |
4036 (73.8)** |
502 (84.8) |
5906 (77.3)** |
154 (90.6) |
6254 (75.9)** |
3076 (79.2) |
4202 (64.7)** |
357 (84.2) |
6921 (69.5)** |
85 (89.5) |
7193 (70.0)** |
| Neoplasms, n(%) | 8832 (47.5) |
1820 (65.7) |
3050 (55.8)** |
389 (65.7) |
4481 (58.6)** |
121 (71.2) |
4749 (58.9)* |
2598 (41.2) |
2364 (36.4)** |
210 (49.5) |
3752 (37.7)** |
50 (52.6) |
3912 (38.0)* |
| Neurological, n(%) | 6827 (36.7) |
1533 (55.4) |
2256 (41.3)** |
350 (59.1) |
3439 (45.0)** |
136 (80.0) |
3653 (45.3)** |
1263 (32.5) |
1775 (27.3)** |
183 (43.2) |
2855 (28.7)** |
70 (73.7) |
2968 (28.9)** |
| Respiratory, n(%) | 9416 (50.6) |
1970 (71.1) |
2844 (52.0)** |
440 (74.3) |
4374 (57.2)** |
135 (79.4) |
4679 (58.0)** |
2060 (53.1) |
2542 (39.1)** |
250 (59.0) |
4352 (43.7)** |
69 (72.6) |
4533 (44.1)** |
| Mental disorder, n(%) | 4784 (25.7) |
1296 (46.8) |
1410 (25.8)** |
347 (58.6) |
2359 (30.9)** |
126 (74.1) |
2580 (32.0)** |
955 (24.6) |
1123 (17.3)** |
179 (42.2) |
1899 (19.1)** |
71 (74.7) |
2007 (19.5)** |
| Mood disorders, n(%) | 2598 (14.0) |
823 (29.7) |
780 (14.3)** |
248 (41.9) |
1355 (17.7)** |
106 (62.4) |
1497 (18.6)** |
484 (12.5) |
511 (7.9)** |
110 (25.9) |
885 (8.9)** |
51 (53.7) |
944 (9.2)** |
p<0.05
p<0.001; Mood disorders is a subset of Mental disorders; AFR=African ancestry; EUR=European ancestry; PMBB=Penn Medicine Biobank; TUD=Tobacco use disorder; AUD=Alcohol use disorder; OUD=Opioid use disorder.
Primary associations of SUD PRS
PRS were calculated for smoking initiation, AUD, and OUD from summary statistics available from GWAS conducted in the Million Veteran Program (Supplemental Table S1). PRS were significantly correlated within each ancestry (Supplemental Table S3). We tested the association of the three PRS with their respective primary phenotypes, using both less stringent and stringent case definitions (Table 2). Using the less stringent case definition, PRSSmoking was strongly positively associated with TUD in both the AFR and EUR samples (AFR cases = 2,769, OR = 1.12, p = 1.4 x 10−8; EUR cases = 3,883, OR = 1.25, p = 3.6 x 10−22). Likewise, the PRSAUD was positively associated with AUD in both the AFR and EUR samples (AFR cases = 592, OR = 1.09, p = 0.04; EUR cases = 424, OR = 1.19, p = 7.1 x 10−4) as was PRSOUD (AFR cases = 170, OR = 1.24, p = 0.01; EUR cases = 95, OR = 1.28, p = 0.02). Similar associations were obtained using the stringent phenotype definition, although PRSOUD was no longer significant in the EUR sample (Table 2).
Table 2.
Polygenic risk scores results by substance use using the PRS-CS method
| Training Set | Testing Set (Penn Medicine Biobank) | ||||||
|---|---|---|---|---|---|---|---|
| SUD | Ancestry | Phenotype [Reference] |
Total Sample Size |
Phenotype | OR | SE | P |
| Tobacco | AFR | Initiation (Contrast I, current vs. never smokers) [10] | 55,890 AFR | Less stringent | 1.12 | 1.06-1.18 | 1.8x10−5 |
| Stringent | 1.12 | 1.07-1.19 | 1.4x10−5 | ||||
| EUR | 209,915 EUR | Less stringent | 1.25 | 1.19-1.31 | 3.6x10−22 | ||
| Stringent | 1.25 | 1.20-1.31 | 5.7x10−22 | ||||
| Alcohol | AFR | AUD [8, 21] | 56,648 AFR | Less stringent | 1.09 | 1.00-1.19 | 0.04 |
| Stringent | 1.11 | 1.01-1.22 | 0.02 | ||||
| EUR | 313,959 EUR | Less stringent | 1.19 | 1.08-1.32 | 7.1x10−4 | ||
| Stringent | 1.21 | 1.09-1.35 | 3.5x10−4 | ||||
| Opioids | AFR | OUD [9] | 31,462 AFR | Less stringent | 1.24 | 1.05-1.47 | 0.01 |
| Stringent | 1.23 | 1.02-1.50 | 0.03 | ||||
| EUR | 80,219 EUR | Less stringent | 1.28 | 1.04-1.58 | 0.02 | ||
| Stringent | 1.23 | 0.97-1.56 | 0.09 | ||||
SUD=Substance use disorder; AFR=African ancestry; EUR=European ancestry. Bolded p-values are p<0.05.
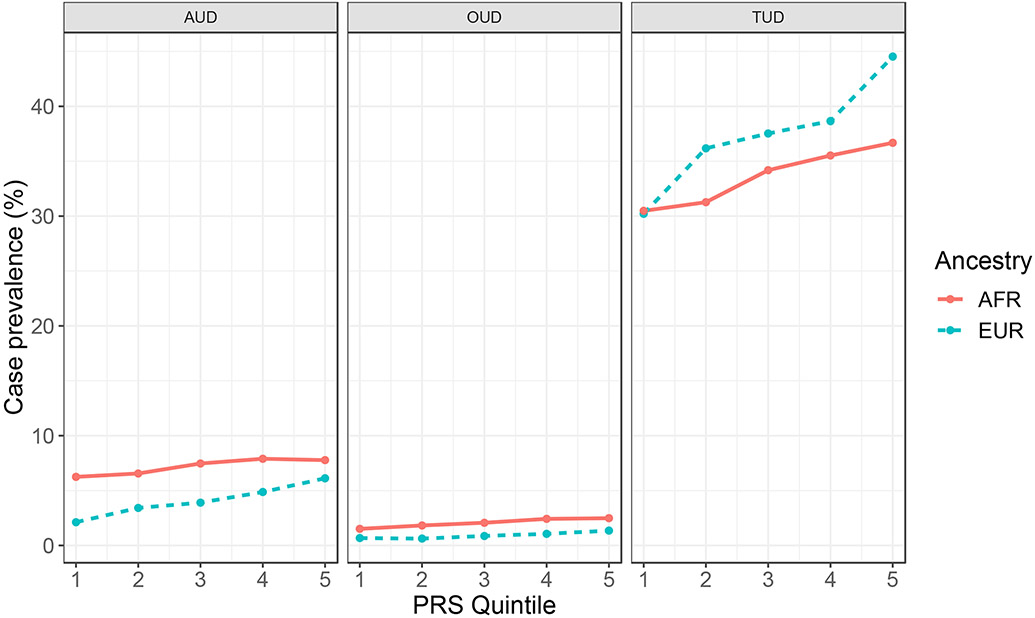
For each phenotype, we determined the case prevalence by PRS quintile. PRSSmoking showed an absolute risk of 36.7% for AFR and 44.5% for EUR in the top quintile (Figure 1), with corresponding odds ratio (OR) of 1.20 (p = 0.004) for AFR and 1.45 (p = 3.6x10−13) (Supplemental Table S4). The top quintile for PRSAUD showed an absolute risk of 7.8% for AFR (OR = 1.13, p = 0.25) and 6.1% for EUR (OR = 1.59, p = 5.2x10−5) (Figure 1; Supplemental Table S4). For PRSOUD, the absolute risk in the top quintile was 2.5% for AFR (OR = 1.28, p = 0.21) and 1.3% for EUR (OR = 1.64, p = 0.03) (Figure 1; Supplemental Table S4).
Figure 1.
Case prevalence (%) by PRS quintile for AUD, OUD, and TUD using the less stringent definition
Phenome-wide analysis of SUD PRS
Tobacco.
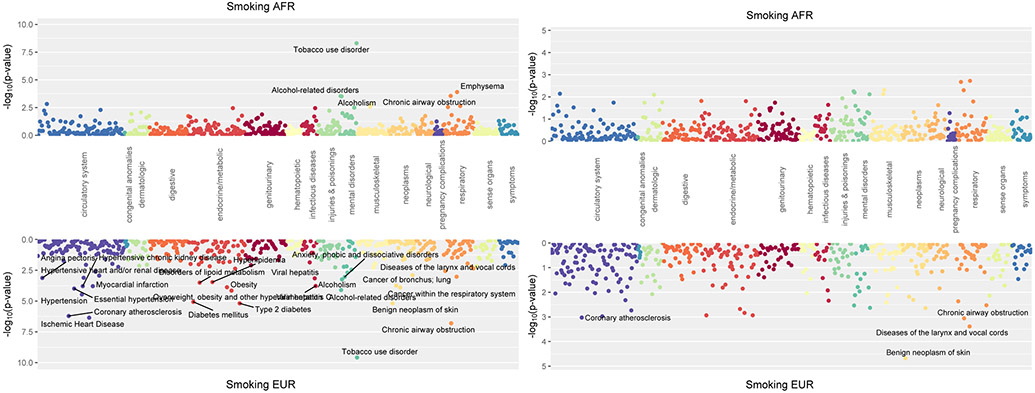
There was a significant association of the AFR PRSSmoking with TUD (OR = 1.19, p = 4.95x10−9) and strong, though after Bonferroni correction not statistically significant, associations with emphysema (OR = 1.34, p = 1.3x10−4), chronic airway obstruction (OR = 1.17, p = 2.8x10−4), alcohol-related disorders (OR = 1.26, p = 2.9x10−4) and alcoholism (OR =1.29, p = 3.0x10−4) (Figure 2A). Covarying for TUD, the AFR PRSSmoking showed no statistically significant associations (Figure 2B). The analysis by sex (Supplemental Figure S1) showed there were strong associations of the PRSSmoking with TUD in both men (OR = 1.23, p= 2.46x10−5) and women (OR = 1.18, p = 3.27x10−5).
Figure 2.
Phenome-wide association of polygenic risk scores for smoking initiation.
A presents the unadjusted smoking initiation results.
B presents PRSSmoking, covarying for TUD.
The EUR PRSSmoking was significantly associated with 12 phenotypes, including increased risk of TUD (OR = 1.25, p = 2.61x10−10), chronic airway obstruction (OR = 1.21, p = 1.57x10−7), ischemic heart disease (OR = 1.14, p = 4.43x10−7), coronary atherosclerosis (OR = 1.14, p = 6.11x10−7) and Type 2 diabetes (OR = 1.14, p = 6.53x10−6) (Figure 2A). Covarying TUD, the only association that remained significant was a protective effect against benign neoplasm of skin (OR = 0.88, p = 2.09x10−5) (Figure 2B). Analysis by sex (Supplemental Figure S1) showed that, in men, PRSSmoking was significantly associated with 10 phenotypes, including TUD (OR = 1.24, p = 1.24x10−7), chronic airway obstruction (OR = 1.22, p = 8.67x10−6), several circulatory system diseases (e.g., hypertension, OR = 1.16, p = 1.57x10−5), and Type 2 diabetes (OR= 1.16, p = 1.87x10−5). In women, PRSSmoking remained strongly associated with TUD (OR = 1.28, p = 7.22x10−5), with smaller associations with thyroid disorders (OR = 0.62, p = 4.8x10−4), benign neoplasm of skin (OR = 0.86, p = 5.3x10−4).
Alcohol.
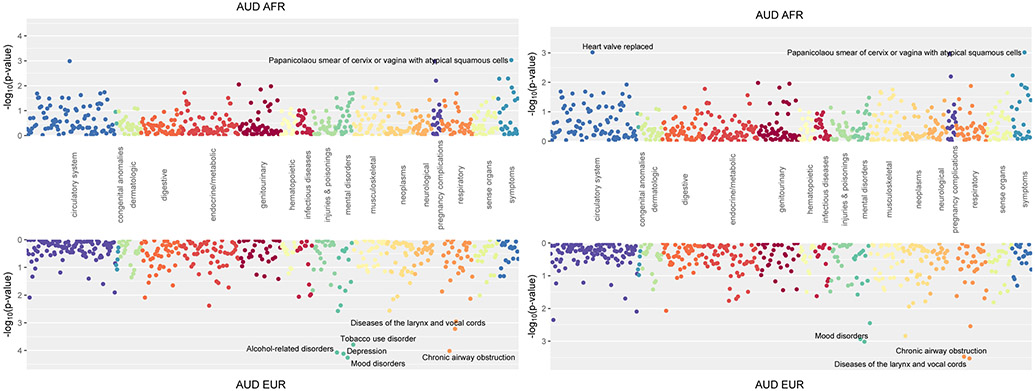
The AFR PRSAUD was not significantly associated with any phenotype (Figure 3A), which was not altered by adding AUD as a covariate (Figure 3B) or examining PRSAUD by sex (Supplemental Figure S2).
Figure 3.
Phenome-wide association of polygenic risk scores for alcohol use disorder (AUD).
A presents the unadjusted AUD results.
B presents PRSAUD, covarying for AUD.
The EUR PRSAUD showed significant positive associations with mood disorders (OR = 1.16, p = 5.49x10−5), depression (OR = 1.16, p = 7.65x10−5), alcohol-related disorders (OR = 1.35, p = 8.49x10−5), and chronic airway obstruction (OR = 1.15, p = 9.63x10−5) (Figure 3A). Further, it was nominally positively associated with TUD (OR = 1.14, p = 1.62x10−4). Covarying AUD demonstrated a nominally significant relationships with protective effect for disease of the larynx and vocal chords (OR = 0.78, p = 2.97x10−4), chronic airway obstruction (OR = 1.13, p = 3.36x10−4) and mood disorders (OR = 1.13, p = 9.62x10−4) (Figure 3B). In men, there were strong positive associations with mood disorders (OR = 1.25, p = 1.12x10−5), depression (OR = 1.25, p = 1.54x10−5), and alcohol-related disorders (OR = 1.43, p = 2.06x10−5), but no strong associations in women (Supplemental Figure S2).
Opioids.
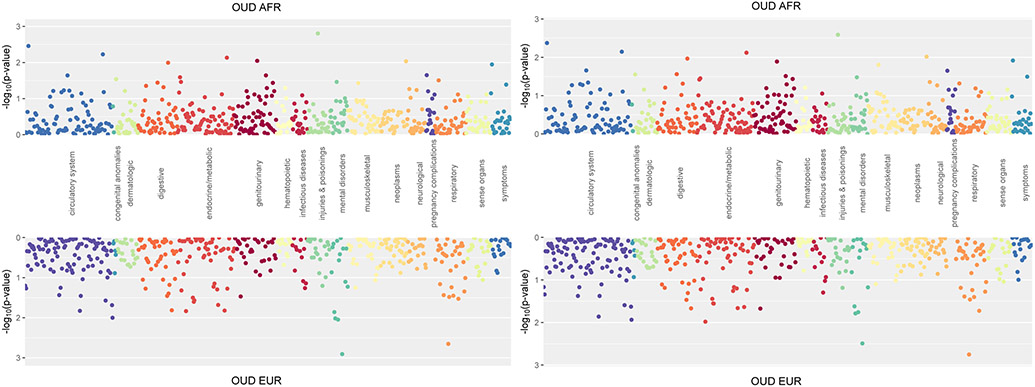
The AFR PRSOUD was not significantly associated with any phenotype (Figure 4A), though there were nominal, positive associations with fracture of ankle and foot (OR = 1.31, p = 1.56x10−3) and acute pulmonary heart disease (OR = 1.22, p = 3.48x10−3). A similar pattern of findings was obtained when OUD was included as a covariate (Figure 4B). Analysis by sex showed no significant associations (Supplemental Figures S3A and S3B).
Figure 4.
Phenome-wide association of polygenic risk scores for opioid use disorder (OUD).
A presents the unadjusted OUD results.
B presents PRSOUD, covarying for OUD.
The EUR PRSOUD was not significantly associated with any phenotype, though there was a nominally positive association with mood disorders (OR = 1.12, p = 1.2x10−3) and diseases of the larynx and vocal chords (OR = 0.82, p = 2.2x10−3) (Figure 4A). Covarying OUD yielded similar results (Figure 4B). When examining PRS by sex, men showed significant positive associations with mood disorders (OR = 1.22, p = 3.89x10−5) and depression (OR = 1.19, p = 3.44x10−4) (Supplemental Figure S3A), while there were no significant associations in women (Supplemental Figure S3B).
Discussion
In this study of 18,612 AFR and EUR individuals from the PMBB, all PRS were associated with their respective diagnoses in both ancestral populations, with the most robust findings being for PRSSmoking. The PRSOUD performed less well than the other SUD PRS. The PheWAS results yielded multiple cross-trait associations which have important implications for our understanding of potential shared genetic etiology for comorbid conditions, such as mood and alcohol use disorders. These findings provide new insights into common genetic factors underlying SUD in both AFR and EUR ancestry samples. Given the increasing attention to the lack of diversity in genetic studies,24 these findings begin to address the gap in our understanding of the genetic risk factor underlying SUD in African Americans.
The strongest associations were found for PRSSmoking, followed by PRSAUD and PRSOUD. The difference in the strength of association between PRS and diagnosis is likely a reflection of both the originating GWAS sample size and the prevalence and accuracy of diagnosis in the PMBB dataset. As in previous studies,25 the variance explained by PRS for all SUDs was small (e.g., a 13% difference in case prevalence between top and bottom quintile for PRSSmoking in EUR individuals). Thus, while indicative of the risk for the respective SUDs, PRS for the substance-related phenotypes of smoking initiation, AUD, and OUD are currently not adequately predictive to identify cases in an unselected sample.
As expected, in addition to its association with TUD in both ancestral samples, PRSSmoking was associated with alcohol-related disorders and other diseases commonly found in smokers, such as coronary and respiratory disorders. These findings align with both the strong phenotypic relationship between smoking and alcohol use and the known genetic overlap between these traits.5,10,26 Interestingly, in both the AFR and EUR samples, these associations were no longer significant after covarying for the primary TUD diagnosis, suggesting that the association between PRS and these phenotypes may be mediated by the TUD diagnosis.
Mirroring known phenotypic comorbidity between AUD and mood disorders,5 the EUR sample PRSAUD was associated with mood disorders and depression. This confirms research in other samples that shows that the PRS for MDD is predictive of alcohol dependence.27 The association between the PRSAUD and chronic airway obstruction is likely a secondary consequence of smoking, which is also nominally positively associated. These findings replicate the previously observed common genetic factors underlying TUD, alcohol-related disorders, mood disorders, and chronic airway obstruction. However, prior PheWASs of alcohol-related PRS in European samples8,21 yielded more significant associations than found here, likely due to their larger sample sizes.
The PRSOUD PheWAS showed no significant associations in either ancestral sample, likely due to the limited power of the PRS as the originating GWAS9 on which they were based was smaller than that of the alcohol8,21 or smoking10 GWAS. In addition, OUD may be underrepresented and under-diagnosed in EHRs in academic medical centers.
There were noteworthy sex differences in our findings, many of which align with established phenotypic patterns regarding SUDs and comorbid conditions. In both men and women, PRSSmoking was significantly associated with TUD. However, there were sex differences in somatic and psychiatric comorbidity; PRSSmoking was not associated with any other mental health conditions in men, whereas it was associated with anxiety disorders in women. This likely reflects the finding that women are more likely to smoke to regulate their mood and mitigate stress28 as well as suffer more stress and anxiety from nicotine withdrawal than men.29 In contrast, PRSSmoking was associated with circulatory system diseases and Type 2 diabetes in men but not women. This could be because men smoke more cigarettes per day, inhale more deeply, and smoke cigarettes with higher nicotine content in men than in women.30
There were also sex differences in the associations with PRSAUD, which were associated with mood disorders in EUR men, but not in either EUR or AFR women or AFR men. Given that the comorbidity of mood disorders and SUDs are typically more common in women than men,31,32 these findings were not expected. The lack of association of the PRSAUD in women could be attributable to the preponderance of males in the MVP GWAS sample that was used to calculate PRS. Data are mixed regarding whether there is a different genetic liability for alcohol-related outcomes between men and women.33,34
The sex-specific association between PRSOUD and mood disorders in EUR men is consistent with findings that mood disorders and OUD are commonly comorbid32 and significantly genetically correlated.9 As with alcohol and tobacco use, there are sex-specific patterns of opioid use, with younger women using smaller amounts of opioids for shorter periods,35 which could either reflect either social or genetic factors.
Limitations of this study include the low prevalence of SUD diagnoses in the PMBB, particularly OUD, which limited statistical power to detect associations. The higher age composition of the PMBB than the general population, associated with a lower prevalence of SUDs, as is typically seen in older adults, likely contributed to the comparatively low prevalence of AUD and OUD. Rates of cardiovascular disease are high in the PMBB because of a concerted effort to recruit from cardiovascular clinics. Therefore, the findings reported here may not generalize to other clinical populations. Additionally, findings from MVP data may not generalize to other populations given the unique nature of the sample (i.e., Veterans, predominantly male, high rates of comorbid conditions). Future studies should consider using as training data summary statistics from GWAS that have covaried for possible influential comorbidities to test whether the associations in this study continue to hold. We were unable to test for sample overlap between the two biobanks, which could bias results; however, this possibility is unlikely given the low percentage of the MVP sample (<2%) that was recruited from the Philadelphia region. Finally, the reliance on EHR diagnoses, rather than those obtained through self-report or diagnostic interviews, may have led to misclassification of disorders and the omission of those that occurred outside of Penn Medicine.
The primary strength of this study is the diversity of the PMBB sample, which enabled us to study the large and growing sample of AFR-ancestry individuals in the PMBB. The availability of summary statistics from independent GWAS of AFR-ancestry individuals enabled us to conduct analyses in this understudied population. For this reason, we limited our selection of GWAS summary statistics to those available from MVP. Larger GWASs for these and related phenotypes are available for European-ancestry samples, but are not yet available for non-European ancestry samples. Another strength of the study is the wealth of phenotypic information available in the Penn Medicine EHR, making it possible to conduct PheWAS. As further GWAS are conducted in diverse ancestral groups, such as those possible in MVP where there are increasing in samples of Latinos and East Asians, and summary statistics become available, we will extend this work to other populations.
This study adds to the growing body of literature demonstrating the genetic liability for SUDs and cross-trait associations with medical and psychiatric comorbidity in AFR- and EUR-ancestry individuals. Identifying differential risk profiles in multiple population groupsand in both sexes may help to interpret the heterogeneity that has challenged our understanding of the genetic architecture of SUD.
Supplementary Material
Funding and Acknowledgements
EEH and HRK are supported by the Crescenz VAMC Mental Illness Research, Education and Clinical Center. RLK is supported in part by NIAAA (K01AA028292) and the Million Veteran Program, Office of Research and Development, Veterans Health Administration awards (I01 CX001734 and I01 BX003341). RLK and AKM are supported in part by the TAPITMAT in Translational Medicine and Therapeutics through NCATS (UL1TR001878).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This publication does not represent the views of the Department of Veterans Affairs or the U.S. government.
We acknowledge and thank the participants of the Penn Medicine Biobank and the Million Veteran Program.
All Regeneron Genetics Center collaborating authors report being employees of Regeneron Pharmaceuticals.
Footnotes
Disclosures: HRK is a member of a Dicerna Pharmaceuticals scientific advisory board, a consultant to Sophrosyne Pharmaceuticals, a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which during the past three years was supported by AbbVie, Alkermes, Dicerna, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor Pharmaceuticals, and Amygdala Neurosciences, Inc., and is named as an inventor on PCT patent application #15/878,640 entitled: "Genotype-guided dosing of opioid agonists," filed January 24, 2018. MDR is on the scientific advisory board for Goldfinch Bio and Cipherome. No other authors have disclosures to report.
Data Availability
GWAS summary statistics were contributed by the Million Veteran Program and downloaded from the dbGAP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v3.p1). The data, analytic methods, and study materials may be made available to other researchers for purposes of reproducing the results or replicating the procedure upon reasonable request to the corresponding author and with the appropriate ethical approval and data sharing agreements.
References
- 1.Substance Abuse and Mental Health Services Administration. Key substance use and mental health indicators in the United States: Results from the 2019 National Survey on Drug Use and Health (HHS Publication No. PEP20-07-01-001, NSDUH Series H-55). Rockville, MD: Center for Behavioral Health Statistics and Quality, Substance Abuse and Mental Health Services Administration. 2020. Retrieved from https://www.samhsa.gov/data/ [Google Scholar]
- 2.Whiteford HA, Ferrari AJ, Degenhardt L, Feigin V, Vos T The global burden of mental, neurological and substance use disorders: an analysis from the Global Burden of Disease Study 2010. PLoS One. 2015;10(2):e0116820. doi: 10.1371/journal.pone.0116820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laramée P, Leonard S, Buchanan-Hughes A, Warnakula S, Daeppen JB, Rehm J. Risk of all-cause mortality in alcohol-dependent individuals: a systematic literature review and meta-analysis. EBioMedicine. 2015;2(10):1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee TG, Rosenheck RA. Association of current and past opioid use disorders with health-related quality of life and employment among US adults. Drug alc depend. 2019;199:122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant BF, Goldstein RB, Saha TD, et al. Epidemiology of DSM-5 alcohol use disorder: results from the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA psychiatry. 2015;72(8):757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant BF, Shmulewitz D, Compton WM. Nicotine use and DSM-IV nicotine dependence in the United States, 2001–2002 and 2012–2013. Am J Psychiatr. 2020;177(11): 1082–1090. [DOI] [PubMed] [Google Scholar]
- 7.Webster LR. Risk factors for opioid-use disorder and overdose. Anesthesia & Analgesia, 2017;125(5):1741–1748. [DOI] [PubMed] [Google Scholar]
- 8.Kranzler HR, Zhou H, Kember RL, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nature Communications. 2019;10:1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H, Rentsch CT, Cheng Z, et al. Association of OPRM1 functional coding variant with opioid use disorder: a genome-wide association study. JAMA psychiatry. 2020A:77(10):1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu K, Li B, McGinnis KA, et al. Genome-wide association study of smoking trajectory and meta-analysis of smoking status in 842,000 individuals. Nature communications. 2020;11(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugrue LP, Desikan RS. What are polygenic scores and why are they important? Jama. 2019;321(18):1820–1821. [DOI] [PubMed] [Google Scholar]
- 12.Inouye M, Abraham G, Nelson CP, et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. Journal of the American College of Cardiology. 2018;72(16):1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Docherty AR, Moscati A, Dick D, et al. Polygenic prediction of the phenome, across ancestry, in emerging adulthood. Psychol Med. 2018;48(11):1814–1823. doi: 10.1017/S0033291717003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kember RL, Merikangas AK, Verma SS, et al. Polygenic risk of psychiatric disorders exhibits cross-trait associations in electronic health record data from European ancestry individuals. Biological Psychiatry. 2020;89(3):236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatoum AS, Johnson EC, Polimanti R, et al. The Addiction Genetic Factor a (g): A Unitary Genetic Vulnerability Characterizes Substance Use Disorders and Their Associations with Common Correlates. medRxiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdellaoui A, Smit DJ, van den Brink W, Denys D, Verweij KJ. Genomic relationships across psychiatric disorders including substance use disorders. Drug Alc Depend. 2021;108535. [DOI] [PubMed] [Google Scholar]
- 17.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26(9):1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nature Genetics. 2016;48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galinsky KJ, Bhatia G, Loh PR, et al. Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. Am J Hum Genet. 2016;98(3):456–472. doi: 10.1016/j.ajhg.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Sealock JM, Sanchez-Roige S, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nature neuroscience. 2020B;23(7):809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge T, Chen CY, Ni Y, et al. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nature Communications. 2019;10:1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denny JC, Bastarache L, Ritchie MD et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell, 2019;177(1):26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilhjálmsson BJ, Yang J, Finucane HK, et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97(4):576–92. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang LH, Whitfield JB, Liu M. et al. Associations between polygenic risk for tobacco and alcohol use and liability to tobacco and alcohol use, and psychiatric disorders in an independent sample of 13,999 Australian adults. Drug alc depend 2019;205:107704. [DOI] [PubMed] [Google Scholar]
- 27.Andersen AM, Pietrzak RH, Kranzler HR, et al. Polygenic scores for major depressive disorder and risk of alcohol dependence. JAMA psychiatry. 2017;74(11):1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cosgrove KP, Wang S, Kim SJ, et al. Sex differences in the brain's dopamine signature of cigarette smoking. J Neurosci. 2014;34(50):16851–5. doi: 10.1523/JNEUROSCI.3661-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres OV, O'Dell LE. Stress is a principal factor that promotes tobacco use in females. Prog Neuropsychopharmacol Biol Psychiatry. 2016;65:260–8. doi: 10.1016/j.pnpbp.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melikian AA, Djordjevic MV, Hosey J, et al. Gender differences relative to smoking behavior and emissions of toxins from mainstream cigarette smoke. Nicotine Tob Res. 2007;9(3):377–87. doi: 10.1080/14622200701188836. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein RB, Dawson DA, Chou SP, Grant BF. Sex differences in prevalence and comorbidity of alcohol and drug use disorders: results from wave 2 of the National Epidemiologic Survey on Alcohol and Related Conditions. J Stud Alc Drugs. 2012;73(6):938–950. 10.15288/jsad.2012.73.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grella CE, Karno MP, Warda US, Niv N, Moore AA. Gender and comorbidity among individuals with opioid use disorders in the NESARC study. Addictive behaviors. 2009;34(6-7):498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salvatore JE, Cho SB, Dick DM. Genes, Environments, and Sex Differences in Alcohol Research. J Stud Alcohol Drugs. 2017;78(4):494–501. doi: 10.15288/jsad.2017.78.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prescott CA, Aggen SH, Kendler KS. Sex differences in the sources of genetic liability to alcohol abuse and dependence in a population-based sample of U.S. twins. Alcohol Clin Exp Res. 1999;23(7):1136–44. doi: 10.1111/j.1530-0277.1999.tb04270.x. [DOI] [PubMed] [Google Scholar]
- 35.Powis B, Griffiths P, Gossop M, Strang J. The differences between male and female drug users: community samples of heroin and cocaine users compared. Subst Use Misuse. 1996;31(5):529–43. doi: 10.3109/10826089609045825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GWAS summary statistics were contributed by the Million Veteran Program and downloaded from the dbGAP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v3.p1). The data, analytic methods, and study materials may be made available to other researchers for purposes of reproducing the results or replicating the procedure upon reasonable request to the corresponding author and with the appropriate ethical approval and data sharing agreements.