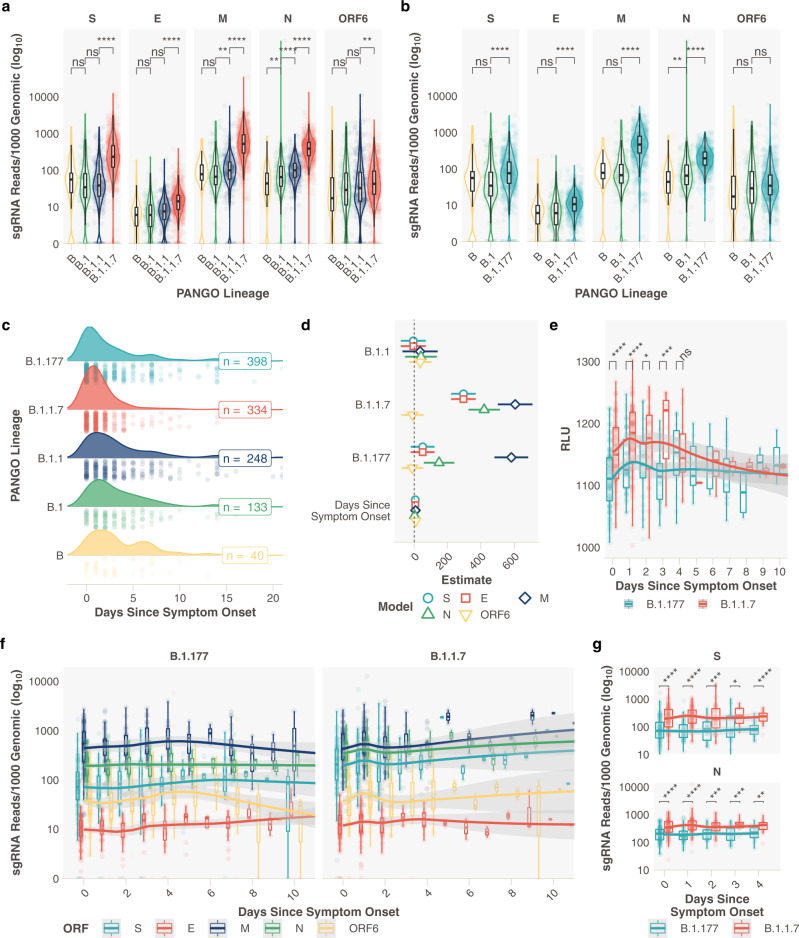
Fig. 2. Relationship between the day of symptom onset at sampling and subgenomic RNA levels in B.1.1.7 and B.1.177 infections.
a Subgenomic RNA data from most abundant ORFs for B.1.1.7 and respective ancestral lineages in Pillar 1 data (Bn = 51, B.1n = 164, B.1.1n = 302, B.1.1.7n = 729). b Subgenomic RNA data from most highly expressed ORFs for B.1.177 and respective ancestral lineages in Pillar 1 data (Bn = 51, B.1n = 164, B.1.177n = 764). c Distribution of the reported days since symptom at the time of sampling for B (n = 40), B.1 (n = 133), B.1.1 (n = 248), B.1.177 (n = 398) and B.1.1.7 (n = 334) lineage infections. d Model estimates and 95% confidence intervals from generalised linear model comparing the effect of lineage and day of symptom onset on sgRNA abundance for most highly abundant ORFs. Reference lineage B.1. No interaction was observed between any lineage and the day of symptom onset. One B.1 sample with outlier N abundance (SHEF-CC370) was excluded. Error bars represent the standard error of the estimate. e Pseudo time course of RLU values for B.1.1.7 and B.1.177, plot restricted to 10 days. (0 Days; B.1.177n = 92, B.1.1.7n = 106. 1 Day; B.1.177n = 52, B.1.1.7n = 102. 2 Days; B.1.177n = 28, B.1.1.7n = 43. 3 Days; B.1.177n = 31, B.1.1.7n = 14. 4 Days; B.1.177n = 17, B.1.1.7n = 19.). f Normalised subgenomic RNA abundance for each of the most abundant ORFs stratified by the reported days since symptom onset and lineage, loess model. The plot is restricted to 10 days. (0 Days; B.1.177n = 115, B.1.1.7n = 93. 1 Day; B.1.177n = 53, B.1.1.7n = 93. 2 Days; B.1.177n = 31, B.1.1.7n = 47. 3 Days; B.1.177n = 31, B.1.1.7n = 12. 4 Days; B.1.177n = 19, B.1.1.7n = 18.). g Normalised sgRNA abundance for N and S is significantly increased at 0-4 days of infection. All p values calculated using an unpaired Wilcoxon signed-rank test, and adjusted for multiple testing with Holm (**** <0.0001, *** <0.001, ** <0.01, * <0.05, ns or blank = not significant). All boxplots depict the 25th, 50th (median), and 75th percentiles, and whiskers represent the most extreme datapoint which is no more than 1.5x the interquartile range. B.1.1.7 is represented in red, B.1.177 in teal, B in yellow, B.1 in green, B.1.1 in navy blue, B.1.1.1 in purple and other lineages in grey apart from D, S subgenomic RNA is teal, E is red, M is navy blue, N in green and ORF6 in yellow.