Fig. 3.
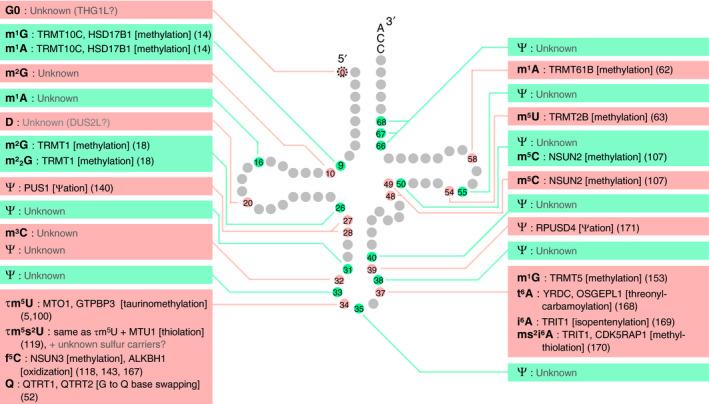
Human mitochondrial (mt) tRNA modifications and modification enzymes. The name of the modification enzyme, the reaction the enzyme is responsible for (in brackets), and the reference (in parentheses) is written next to the species of tRNA modification [52]. Note that the strength of evidence varies between different studies, ranging from checking only that the protein is necessary for modification to fully confirming that the protein is both necessary and sufficient for the modification. For the structures of modifications not depicted in Fig. 1C, please refer to the RNA Modification Database (https://mods.rna.albany.edu). The secondary structures of many mt tRNAs are different from the canonical cloverleaf structure in three ways [13, 46, 224, 225, 226, 227]: (a) mt tRNASer(AGY) lacks the entire D loop, (b) mt tRNASer(UCN) lacks U8 and has a small D loop, a small variable loop, and an extended anticodon stem, and (c) several mt tRNAs do not have canonical D loop/T loop interactions and instead have alternative interactions. Abbreviations not described in Figs. 1 and 2: τm5s2U (5‐taurinomethyl‐2‐thiouridine) and ms2i6A (2‐methylthio‐N6‐isopentenyladenosine).
