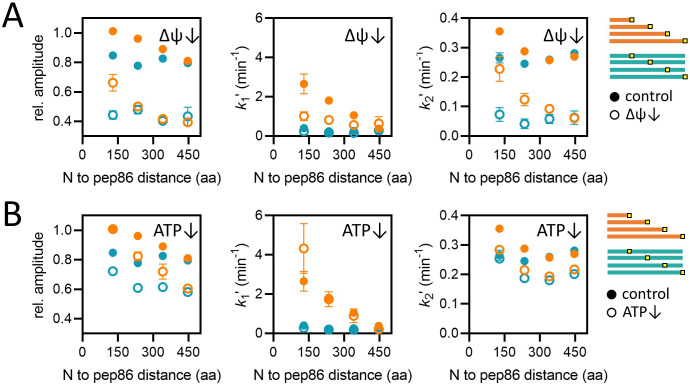
Figure 4. Effects of energy depletion on import of the length and position variants.
(A) Import in the presence (solid circles) or absence (open circles) of ∆ψ, for the length (orange) and position (teal) series. Depletion of ∆ψ was achieved by a 5-min pre-treatment of mitochondria with 10 nM valinomycin. Plots show amplitude (left), k1′ (middle), and k2′ (right) extracted from two-step fits to import traces as a function of PCP length or pep86 position. Each point is the average and SEM of three independent biological replicates. (B) As in panel A, but without (solid circles) or with (open circles) ATP depletion instead of valinomycin. Matrix ATP was depleted by excluding ATP and its regenerating system from the assay mix (see Results section for full description).