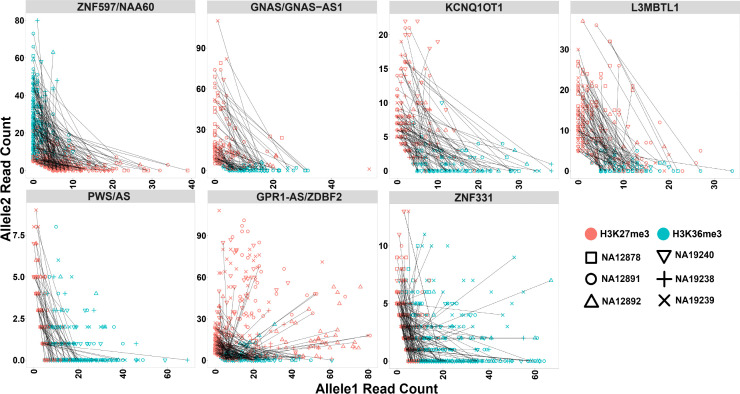
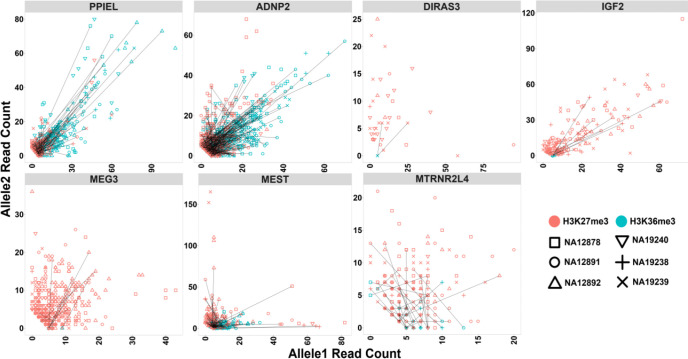
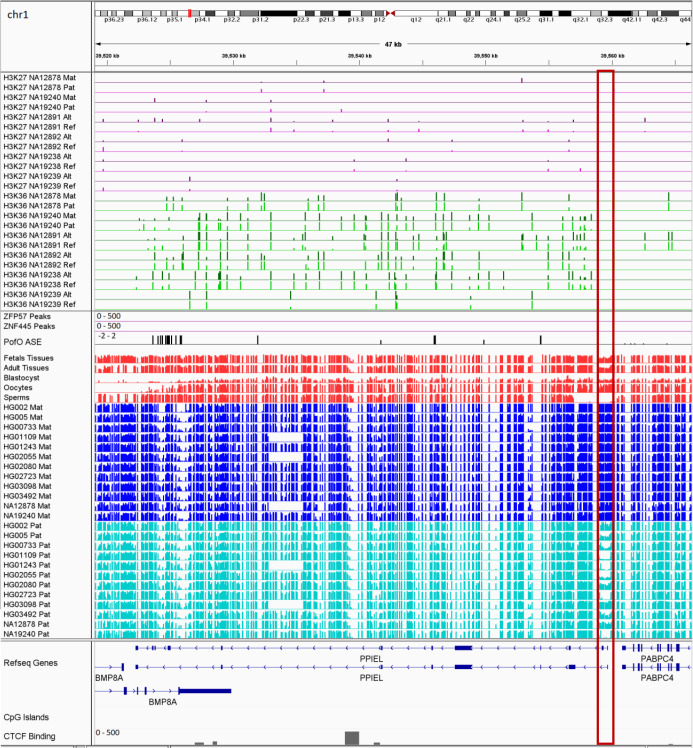
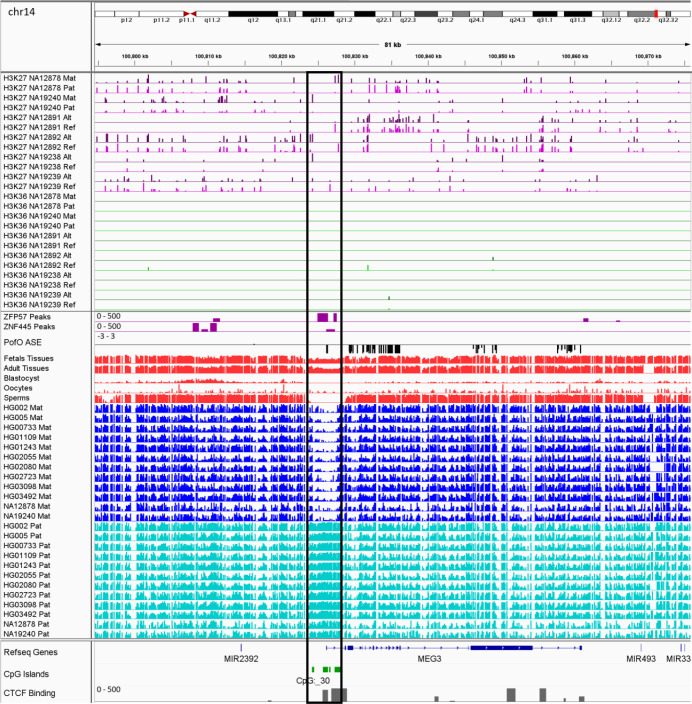
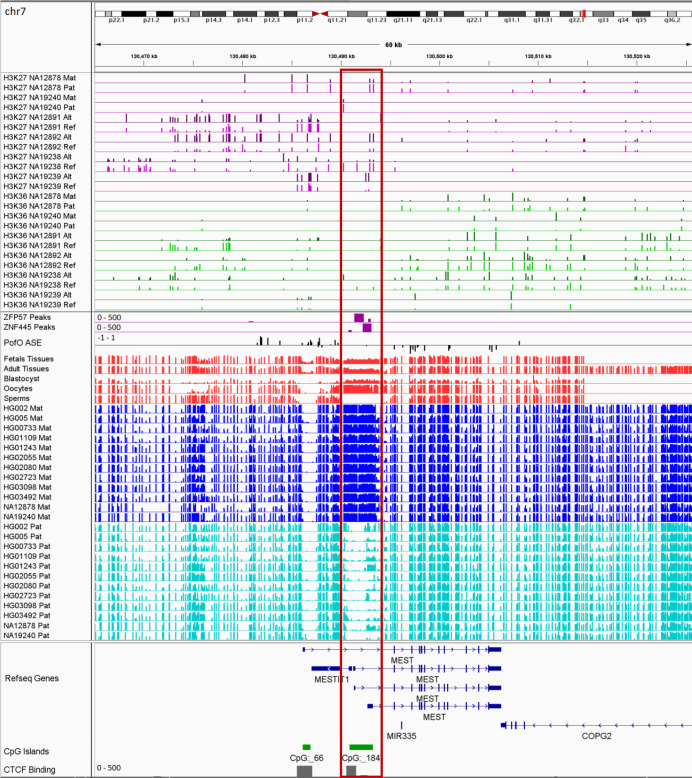
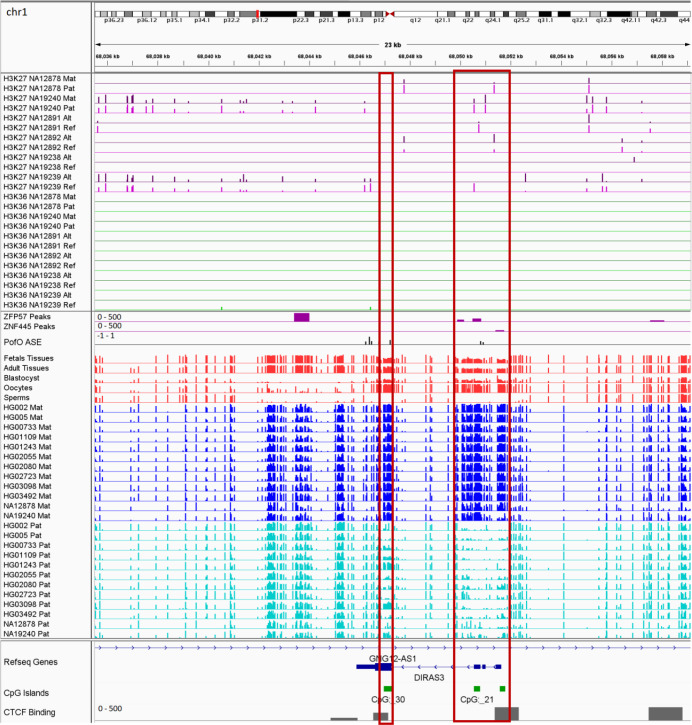
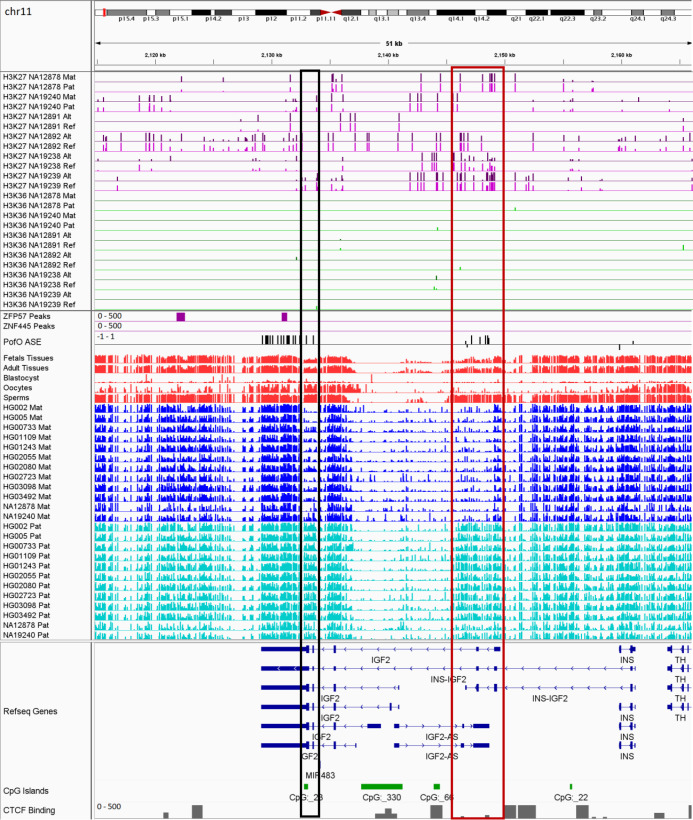
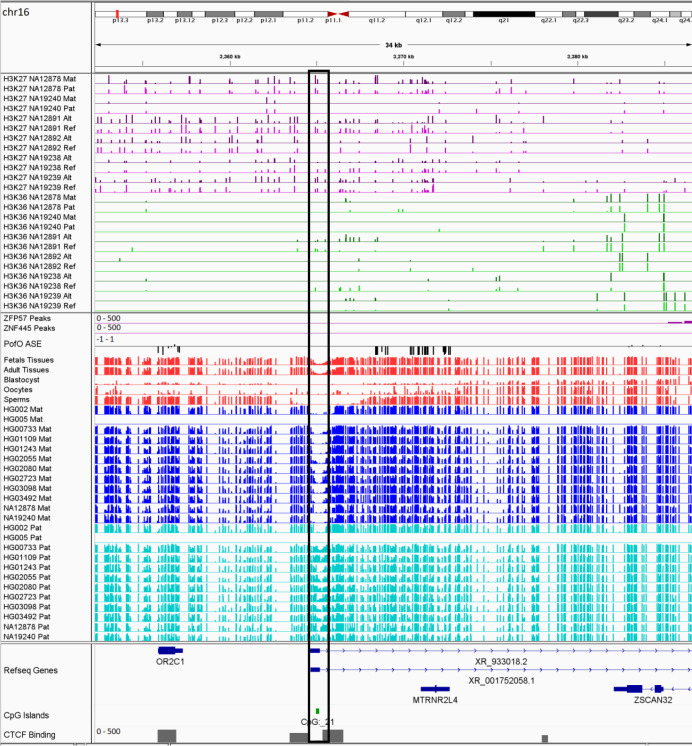
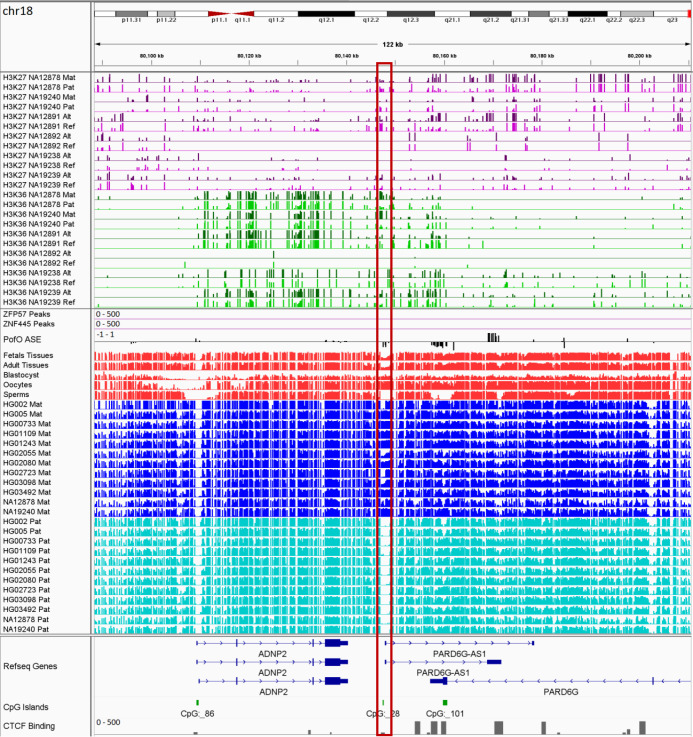
Figure 9. Mutual exclusive allelic H3K36me3 and H3K27me3 histone marks at seven parent-of-origin (PofO) methylation-biased blocks.
All blocks demonstrate allelic H3K36me3 on hypermethylated allele and H3K27me3 on hypomethylated allele. For NA12878 and NA19240, allele1 is the paternal and allele2 is maternal. For sake of visualization in other four cell lines without parental information, allele1 for H3K36me3 mark demonstrates the allele with more mapped reads at all blocks except ZNF597/NAA60. Therefore, for H3K36me3 we swapped the reference allele read count with the alternative allele read count if the reference allele count was less than the alternative allele count. At ZNF597/NAA60, we swapped the reference allele read count with the alternative allele read count if the reference had higher read count. We also swapped the reference and the alternative allele counts for the same SNVs for H3K27me3. Each point represents a heterozygous SNV. Lines are connecting SNVs that have mapped reads for both histone modifications.