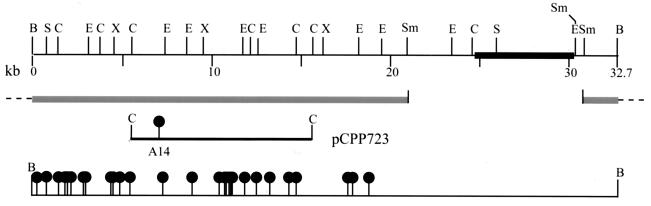
FIG. 4.
Physical map of pCPP719 (top), with the DNA of the pCPP9 vector indicated as a thick bar. The ClaI fragment resulting from a partial digest of pCPP810, containing the mini-Tn5Cm insertion A14 (bottom) was cloned into pBR322 to create pCPP723. This clone was used for marker exchange of insertion A14 into Eh318 and Eh421 (PanA−) to generate Eh439 (PanB−) and Eh440 (PanAB−), respectively. At the bottom is a map of the locations of mini-Tn5Cm insertions (filled lollipops), all of which resulted in abolished pantocin B production. The 23-kb SmaI fragment that was radioactively labeled for use in Southern blot experiments is indicated as two solid gray bars (below map). The dashed lines indicate the points of continuation in the circular pCPP719, and tick marks indicate the points of SmaI cleavage. Abbreviations: B, BamHI; C, ClaI; E, EcoRI; S, SalII; Sm, SmaI; X, XbaI. The figure was generated from Innovative Data Design MacDraft, converted to MacroMedia FreeHand 8, and printed from Adobe Photoshop 5.5.