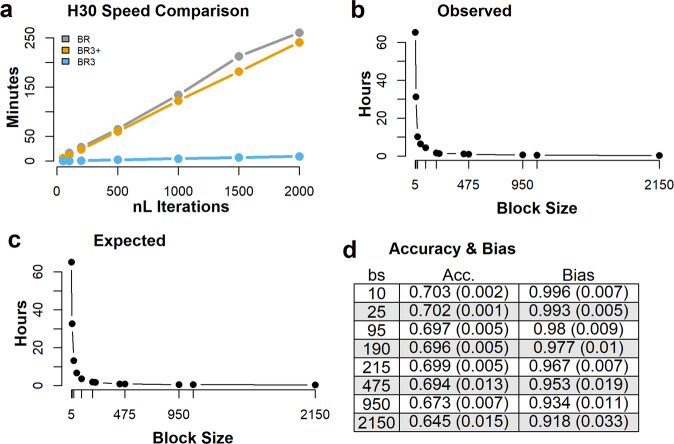
Fig. 5. Processing speeds for the simulated data sets.
a Y-axis is time in minutes to process 20,000 phenotypic records of 400,000 SNPs for the 3 Bayes R configurations as specified in Fig. 1. b Computing time in hours of BR3 with respect to changing block size for Markov chain lengths of 10,000 and for block sizes: , for the simulated data set using 41,925 phenotype records and 400,000 SNPs composed of Holsteins and Jersey cows only, c ratio of , for the same block sizes, n, and where nR is the number of records. Note plot in panel c is scaled to have the same range (min and max) given in plot b. d Aussie Reds genomic prediction mean accuracies (Acc.) and biases (standard deviations) from 5 MCMC chains of length 10,000 each, for selected block sizes associated to the timings given in b.