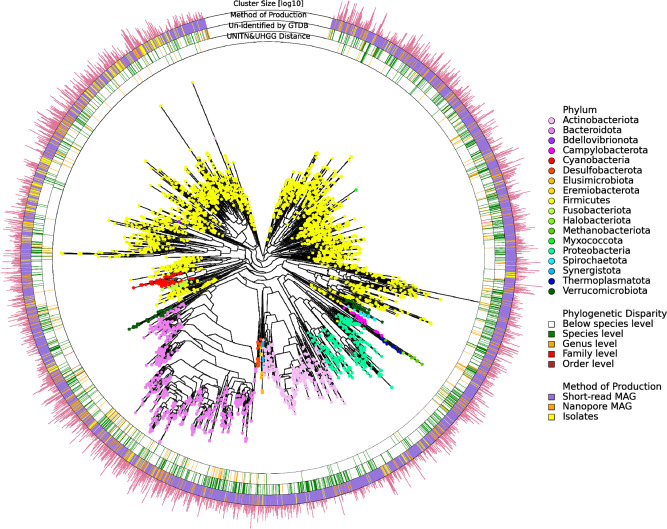
Fig. 1. Phylogenetic tree of the new human gut microbiome genome reference set.
Inside the circle is a phylogenetic tree of the WIS representative genomes colored according to their Genome Taxonomy Database (GTDB) assigned phylum (see color legend). Inner ring shows the distance each of the WIS genomes has from the closest genome in the UNITN or UHGG reference sets, stratified by its phylogenetic distance level—below species level in white, species level in green, genus level in orange, family level in red and order level in brown. Second ring (counting from the inside) shows the highest phylogenetic level the genome was un-identified at, by GTDB—same colors as in the inner ring. Third ring shows the genome production method—short-read metagenome-assembled genomes (MAG) in purple, nanopore MAG in orange, and isolates in yellow. Outer ring line length is proportional to the log10 of the cluster size each genome represents.