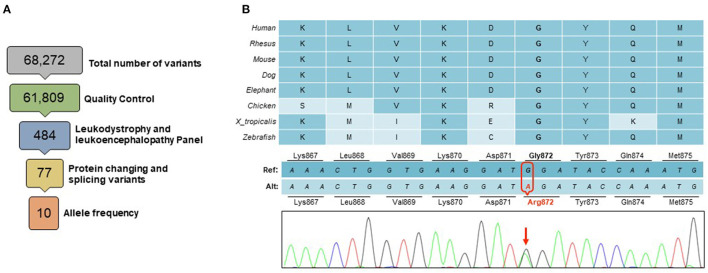
Figure 2.
Genetic findings. (A) A total of 68,272 variants were identified within the targeted exome. Variants of low quality were removed through quality control (GQ <20, RD <10, AF <0.25). Among a panel of 165 genes known to be associated with leukodystrophies and leukoencephalopathies, a total of 484 variants were identified, of which 77 were protein changing or splicing variants. Only 10 of these had an allele frequency <0.10 in gnomAD database, including a novel heterozygous missense variant in the CSF1R gene (B) This change in a highly conserved amino acid in the CSF1R tyrosine domain (p.Gly872Arg), was predicted to be deleterious by multiple in silico tools.