Abstract
We determined 36 310 bovine expressed sequence tag (EST) sequences using 10 different cDNA libraries. For massive EST sequencing, we devised a new system with two major features. First, we constructed cDNA libraries in which the poly(A) tails were removed using nested deletion at the 3′-ends. This permitted high quality reading of sequences from the 3′-end of the cDNA, which is otherwise difficult to do. Second, we increased throughput by sequencing directly on templates generated by colony PCR. Using this system, we determined 600 cDNA sequences per day. The read-out length was >450 bases in >90% of the sequences. Furthermore, we established a data management system for analyses, storage and manipulation of the sequence data. Finally, 16 358 non-redundant ESTs were derived from ∼6900 independent genes. These data will facilitate construction of a precise comparative map across mammalian species and isolate the functional genes that govern economic traits. This system is applicable to other organisms, including livestock, for which EST data are limited.
INTRODUCTION
Single pass sequencing of cDNAs to generate expressed sequence tags (ESTs) is an established area of genome studies. Randomly determined cDNA sequences attract considerable interest as a catalog of expressed genes from a given organism. EST sequences are highly conserved between mammals, therefore a dense gene map will be a powerful tool to construct a precise comparative map (1). In human, 3 400 000 EST sequences have been deposited in a public database and over 30 000 genes have been physically mapped (2). In addition, the draft sequence of the human genome has recently become available (3,4). Therefore, the map position of any gene is available by direct comparison of cDNA and genomic sequences. In mouse, 1 960 000 EST sequences have been deposited in a public database. Approximately 3900 orthologous pairs between human and mouse were recently identified in which ∼3000 genes were mapped in the mouse genome and ∼180 conserved segments were defined (3). Thus, extensive comparison of the mapping data is indispensable to make use of the wealth of data generated by the Human and Mouse Genome Projects for the isolation of the genes underlying important traits in livestock (5–7).
Economic trait loci (ETL) controlling traits such as growth, fatness and milk yield were recently mapped in livestock (8–10). In addition, recent work demonstrated that a common region between cattle and mouse regulates a quantitative trait such as tolerance to trypanosome infection (11). Therefore, a precise comparative map enables us to use not only livestock but also model animals such as mouse to discover ETL genes. In domestic species, however, the numbers of ESTs deposited in the public databases are quite limited. This limitation has impeded the progress of both constructing a precise comparative map and the activity of candidate positional cloning. Recently, an ordered comparative map between cattle and human including 638 orthlogs was reported (12), but more data are required to further refine conserved segments.
To construct a precise comparative map, radiation hybrid (RH) mapping is a convenient method for gene mapping because the markers need not be polymorphic, and thus large-scale mapping is easier. For RH mapping of the genes, it is desirable to design PCR primers in the non-coding region (3′-UTR) to prevent amplification of host DNA present in the hybrid cells (2,13). A large number of bovine ESTs that were recently deposited in a public database by others (14), however, lack information regarding the 3′-end of the cDNA. Furthermore, the 3′-UTR information is useful for clustering a large number of ESTs, being especially effective for discrimination of gene families.
We report the determination of 36 310 bovine ESTs that are mostly composed of a pair of 5′- and 3′-sequences of a cDNA clone, using newly constructed poly(A) tail-removed cDNA libraries. These ESTs will be a valuable resource for livestock ESTs and as markers for RH mapping to construct a precise comparative map.
MATERIALS AND METHODS
Preparation of cDNA libraries
Total RNA was extracted using Trizol (Gibco BRL, Rockville, MD) from several fetal tissues of Holstein cattle, including kidney, liver, rumen, brain, cartilage, lung and ovary, as well as from an adipocyte cell line established from Japanese Black cattle (15). Poly(A)+ RNA was isolated using oligotex-dT30 (Takara, Japan). cDNA synthesis and library construction were performed using the SuperScript λZipLox system (Gibco BRL). In each library, 3 × 105–10 × 105 independent phage transfectants were obtained. Each library was amplified and transfected into Escherichia coli DH10B Zip to convert it to a plasmid library by Cre/loxP-mediated in vivo excision. cDNA clones were pooled and plasmid DNA was extracted from each library. The plasmid was cleaved using the NotI restriction enzyme, followed by nested deletion to remove the poly(A) tail using a Nested Deletion Kit (Amersham Pharmacia Biotech, UK). Deletion conditions were optimized for appropriate deletion sizes (50–100 bp). Briefly, we maintained strict amounts of the linear DNA (1 µg) and time for deletion (1 min). After self-ligation, DNA was transformed into ElectroMax (Gibco BRL) based on the manufacturer’s recommendations. Colonies were picked for subsequent PCR amplification and sequencing.
Preparation of sequencing templates
Each colony was subjected to colony PCR and culture. For colony PCR and DNA sequencing we used the primers CGTCCCATTCGCCATTCAG for the 5′-end and GCTCTAATACGACTCACTATAGGG for the 3′-end. The primer for the 3′-end was located 210 bp downstream of the NotI site in the vector to ensure reliable PCR amplification after deletion. Briefly, a small amount of the colony was suspended in 50 µl of the reaction mixture containing 1× regular PCR buffer consisting of 0.2 µM each primer, 50 µM dNTP, and 2.5 U Taq DNA polymerase (Takara, Japan). The PCR consisted of 35 cycles of 95°C for 30 s for denaturing, 55°C for 30 s for annealing, 72°C for 3 min for extension. After PCR amplification, the products were analyzed using 1% agarose gel electrophoresis, and 1 µl of the PCR product that produced the appropriate length and signal was used directly in a DNA sequencing reaction.
DNA sequencing
For the clones derived from ordinary cDNA libraries [i.e. with poly(A) tails], plasmid DNA was extracted as usual and the sequencing reaction was performed using a BigDye primer kit (P.E. Biosystems, Japan). Otherwise, the sequencing reaction was performed in 10 µl of a reaction mixture containing 6 µl of water, 4 µl of BigDye terminator (P.E. Biosystems), 0.2 µM sequence-specific primer and 1 µl of the PCR product. The proportion of PCR product and conditions for sequencing were crucial for a robust sequence signal without purifying the PCR fragments as template DNA. Cycle sequencing was performed under the conditions of 35 cycles of 95°C for 30 s for denaturing, 55°C for 30 s for annealing, 60°C for 4 min for extension. Sequencing was performed on an ABI 3700 Capillary Sequencer. The quality of the sequence was assessed using Phred software (16,17).
Data management system
A series of sequence data processing procedures were automatically performed using an in-house system, named cDNA Information Manager. The cDNA Information Manager is composed of a sequence database and five servers: FTP and HTTP servers, Sequence Processor, Sequence Analyzer and Sequence Manager. The sequence database is used to store sequence data and analysis reports, etc. The FTP server is used to enter new sequences from a Macintosh computer to this system. The Sequence Processor performs basic processing of new sequences, including trimming sequence ends, quality checking and clustering. The Sequence Analyzer performs homology searches against the GenBank database and designs PCR primers targeting the 3′-ESTs from the sequences. The Sequence Manager provides data from the database at the user’s request using the HTTP server. The cDNA Information Manager allows direct access to the Human Gene Map (http://w ww.ncbi.nlm.nih.gov/genemap/) and Unigene (http://www.ncbi.nlm.nih.gov/UniGene/) databases (NCBI).
Sequence data processing and clustering
EST sequences are submitted to the data management system, cDNA Information Manager. The ambiguous regions are removed from the ends of the sequences to achieve less than one ambiguity out of 20 bases in the text files. Then, the vector sequence was searched using BlastN v.2.0.9 (18) and removed. Sequences that were <100 nt long or had >7% ambiguity were removed from the data flow. After basic processing, the sequences were clustered among previously generated ESTs based on sequence identity using BlastN. The criteria were more than 300 bits (for conventional cDNA libraries) and 400 bits [for poly(A) tail-removed cDNA libraries] of the Blast score with 80% nucleotide identity. An ID was allocated to the sequences that had no similarity to earlier entries in our ESTs, followed by homology searches against the GenBank database.
Accession numbers
Bovine ESTs described in this paper were deposited in the DDBJ database, accession nos AV588548–AV618892 and AV662325–AV668289.
RESULTS AND DISCUSSION
Establishment of a high throughput EST sequencing system
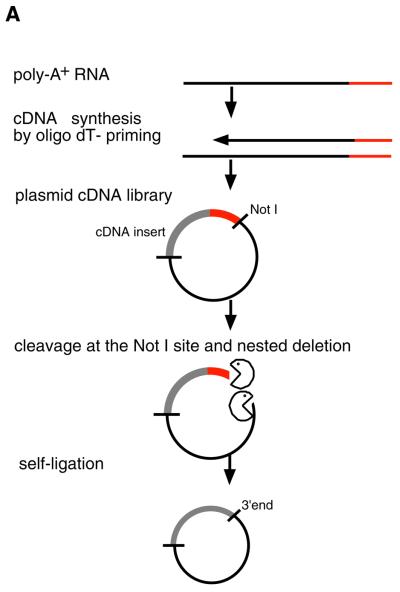
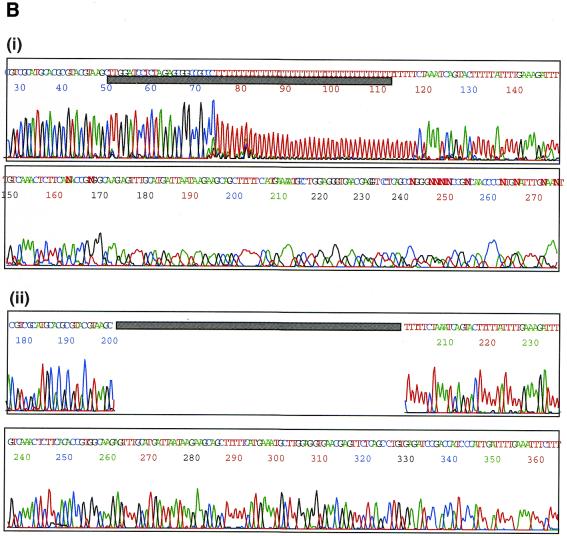
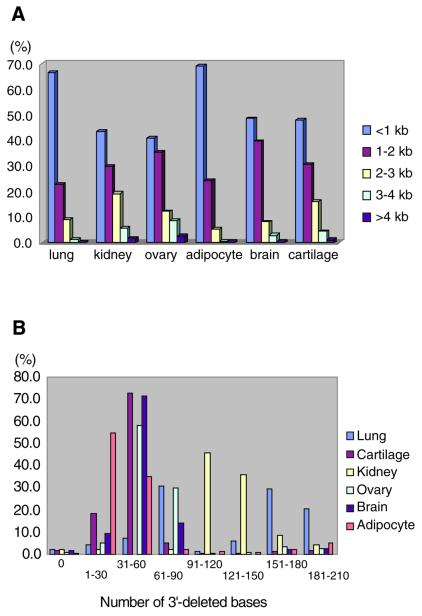
The basic principle for preparation of a poly(A) tail-removed cDNA library is schematically presented in Figure 1A. Bacterial colonies of the cDNA clones were directly subjected to PCR amplification. The 3′ priming site was located 210 bp downstream of the NotI site to ensure that it is out of the range of deletion, because the exonuclease excises nucleotides in both directions from the NotI site. We confirmed that >60% of the cDNA clones were successfully amplified. Direct sequencing of colony PCR products was performed without prior purification. Removal of the poly(A) tail clearly improved the quality of the 3′-sequences (Fig. 1B). According to the evaluation by Phred software (16,17), base calling was possible beyond 450 bp in 90% of the cases (data not shown). Thus, we constructed six poly(A) tail-removed cDNA libraries from various tissues. The size of the cDNA inserts in each library ranged from 0.7 to 4 kb, the average size being 1.5 kb (Fig. 2A).
Figure 1.
(A) The flow chart for preparation of a poly(A) tail-removed cDNA library. An oligo(dT)-primed cDNA library was constructed by a conventional method. Plasmid DNA was extracted from pooled cDNA clones and cleaved at the NotI site located at the cDNA 3′-terminus. The DNA fragments were digested with exonuclease III and mung bean nuclease to remove the poly(A) tail (shown as a red line), followed by self-ligation. Escherichia coli was transformed with the DNA, resulting in a poly(A) tail-removed cDNA library. cDNA inserts were amplified by colony PCR and used as templates for DNA sequencing. (B) Comparison of the sequences from the 3′-ends of the cDNAs. The sequences were derived from bovine osteonectin cDNA clones with (i) and without (ii) a poly(A) tail. Removal of the poly(A) tail clearly improved sequence quality. Shadowed bars correspond to the deleted region.
Figure 2.
(A) Length of cDNA inserts. cDNA inserts were amplified by PCR and used as templates for DNA sequencing. The data for 1920 cDNA clones in each library are shown. (B) Length of deletion from the 3′-end of the cDNA. The size of the deletion was calculated as the number of excised nucleotides from the NotI site to the insert–vector junction.
The size of the deletion as calculated from the distance between the NotI site and the insert–vector junction is shown in Figure 2B. The size of the deletion was within 60 bp in 80% of the clones in each library, except for the lung cDNA library, which was composed of two preparations. In the cDNA libraries, the average deletion sizes were 41 (adipocyte), 45 (cartilage), 52 (brain), 61 (ovary), 121 (kidney) and 125 bp (lung). The procedure was thus deemed sufficient to remove the poly(A) tail while leaving much of the 3′-UTR.
Construction of a data management system
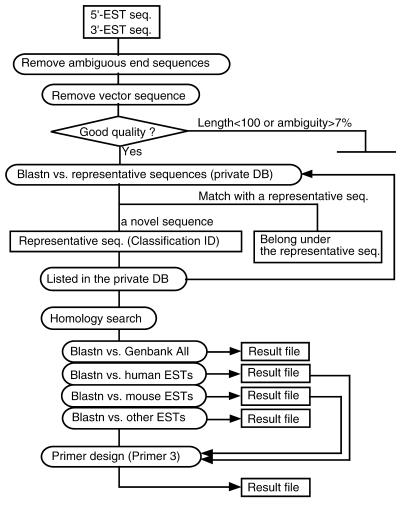
To deal with a large amount of sequence information, we established a data management system, called the cDNA Information Manager. The flow chart of the data analysis is shown in Figure 3. The sequences were subjected to basic processing, including trimming of ambiguous ends and vector sequence, checking sequence quality and clustering based on identity. Although dispersed repeats in ESTs were not masked out, errors in clustering due to dispersed repeats occurred in <1% of sequences (data not shown). A new ID number was allocated to novel sequences, followed by homology searches against the GenBank database. PCR primers were automatically designed for the 3′-sequences for mapping.
Figure 3.
(A) The structure of the data management system, cDNA Information Manager. Text files of the EST sequences produced from a sequencer are submitted to cDNA Information Manager. The sequences are processed and clustered among the sequences deposited in our database. Then novel sequences are subjected to homology searches against each division of the GenBank database, followed by primer design as for the 3′-ESTs. In order to design bovine-specific primers, the results of the homology searches against human and mouse ESTs were linked to Primer 3 software (19).
The results of the analyses, such as homology searches and primer design, can be browsed and the data can be manipulated as necessary. Our data management system is linked to the Human Gene Map (http://www.ncbi.nlm.nih.gov/genemap/) and Unigene (http://www.ncbi.nlm.nih.gov/UniGene/) databases (NCBI) based on homology to human ESTs. In addition, we can obtain various statistics, for example redundancy and the results of homology searches, using custom made programs.
Characterization of ESTs
We generated 19 500 sequences from the 5′-end and 16 800 sequences from the 3′-end of cDNAs. These were clustered into 16 400 groups, in which 8600 and 7800 IDs were allocated to 5′ - and 3′-ESTs, respectively (Table 1). Approximately 12% of the IDs included both 5′- and 3′-ESTs in a group due to short cDNA inserts.
Table 1. Number of ESTs in each library.
| cDNA library | No. of determined sequences | No. of unique ESTs | ||
|---|---|---|---|---|
| 5′ | 3′ | 5′ | 3′ | |
| Adipocyte | 496 | 251 | 380 | 214 |
| Kidney | 313 | 295 | 272 | 262 |
| Liver | 232 | 91 | 125 | 62 |
| Rumen | 380 | 168 | 283 | 140 |
| Adipocyte [poly(A)-removed] | 2016 | 1632 | 765 | 759 |
| Brain [poly(A)-removed] | 3841 | 3471 | 1396 | 1525 |
| Cartilage [poly(A)-removed] | 4147 | 3764 | 1684 | 1609 |
| Kidney [poly(A)-removed] | 4167 | 3772 | 1962 | 1730 |
| Lung [poly(A)-removed] | 2175 | 1866 | 1016 | 874 |
| Ovary [poly(A)-removed] | 1730 | 1503 | 673 | 627 |
| Total | 19497 | 16813 | 8556 | 7802 |
| 36310 | 16358 | |||
To confirm that the deletion from the 3′-end did not affect EST clustering, we estimated the overlap of the 3′-ESTs to which IDs were allocated, by examining the number of IDs with high homology (score ≥ 400 bits, E < e – 109) to the same gene. Thus, ∼90% of 1500 3′-ESTs matched unique genes in the GenBank database. This suggests that interference with EST clustering due to deletion was negligible and the criteria for clustering were appropriate. Thus, we estimate that the ESTs represent ∼6900 genes.
Next, clone redundancy was analyzed as the number of 3′-ESTs included in a cluster (Table 2). All 3′-ESTs (16 813) were clustered into 8331 groups, 529 of which had IDs allocated to 5′-ESTs. Seventy-five percent of the groups included a single 3′-EST, and those unique sequences represented 37% of the overall 3′-sequences. The groups with redundancy of less than five times accounted for >95% of the total groups and corresponded to 64% of the overall 3′-sequences. Therefore, our massive sequencing was effective for isolation of new genes. Some highly redundant sequences were observed (Table 3); most of them were housekeeping genes such as elongation factors, extracellular matrix proteins and ribosomal proteins. A sequence showing a redundancy of 118 times could not be identified from the homology search, but had very weak similarity to human mRNA for insulin-like growth factor II (score = 52 bits, E = 1e – 05), as well as its 5′-sequence. There was also a high frequency of the mitochondrial genome, constituting 0.6% of the 3′-sequences.
Table 2. Redundancy of 3′-ESTs.
| Redundancya | No. of groupsb | Ratio to total groups (%) | Ratio to overall 3′-sequences (%) |
|---|---|---|---|
| 1 | 6269 | 75.2 | 37.3 |
| 2 | 1016 | 12.2 | 12.1 |
| 3 | 380 | 4.6 | 6.8 |
| 4 | 202 | 2.4 | 4.8 |
| 5 | 97 | 1.2 | 2.9 |
| 6–10 | 208 | 2.5 | 9.2 |
| 11–50 | 143 | 1.7 | 17.0 |
| 50–100 | 10 | 0.1 | 3.8 |
| >100 | 6 | 0.1 | 6.3 |
| Total | 8331 | 100.0 | 100.0 |
aThe number of 3′-ESTs included in a group to which an ID was allocated.
bThe number of groups that include a given number of 3′-ESTs.
Table 3. List of highly redundant cDNA clones.
| Redundancya | No. of groupsb | Gene product or gene name |
|---|---|---|
| 51 | 1 | Ribosomal protein L23 |
| 54 | 2 | Ribosomal protein L7a; acidic ribosomal phosphoprotein P1 |
| 57 | 3 | Bone proteoglycan II; ribosomal protein L3; ribosomal protein L4 |
| 70 | 2 | Gβ-like protein; acidic ribosomal phosphoprotein PO |
| 72 | 1 | Glyceraldehyde 3-phosphate dehydrogenase |
| 94 | 1 | Pro-α1 (I) chain of type I procollagen |
| 105 | 1 | Mitochondrial genome |
| 118 | 1 | Weak homology to human mRNA for insulin-like growth factor II |
| 119 | 1 | 60S ribosomal protein L4 |
| 175 | 1 | Elongation factor 1γ |
| 176 | 1 | C10 protein (laminin receptor) |
| 362 | 1 | Elongation factor 1α |
The results of the homology searches against each division of the GenBank database are shown in Table 4. Scores of 400 and 600 bits were used as a threshold against bovine ESTs, while 200 and 400 bits were used against human and mouse ESTs in order to eliminate possible effects of dispersed repeats, because dispersed repeats usually show less than 400 and 150 bits of the scores against bovine and other ESTs, respectively. More than 50% of the ESTs were novel bovine sequences, providing valuable data. Forty-five percent of the 3′-ESTs were thought to be identical to the 5′-sequences of bovine ESTs deposited in a public database, suggesting that they had been derived from short cDNAs. Homology to human ESTs had a higher score than to mouse ESTs, which is consistent with a previous report that rodents have undergone more sequence changes than other mammals (20). In general, the translated region of cDNA is more conserved than the non-coding region among species. As expected, the homology of the 5′-ESTs to mouse or human ESTs was significantly higher than that of the 3′-ESTs (Table 4), which was thought to reflect the fact that the 5′-sequences contained coding regions. Thus the 5′-ESTs were useful for identifying gene functions, while the 3′-ESTs, which were less conserved among species, were useful for designing bovine-specific primers for RH mapping.
Table 4. Results of the homology searches against a public EST database.
| 5′-EST, 8556 clones |
3′-EST, 7802 clones |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| >200 bits | >400 bits | >600 bits | >200 bits | >400 bits | >600 bits | |||||||
| (n) | (%) | (n) | (%) | (n) | (%) | (n) | (%) | (n) | (%) | (n) | (%) | |
| ND, not determined | ||||||||||||
| Versus bovine ESTsa | ND | ND | 4137 | 48.4 | 2941 | 34.4 | ND | ND | 3507 | 45.0 | 2224 | 28.5 |
| Versus human ESTsb | 4391 | 51.3 | 2554 | 29.9 | ND | ND | 2937 | 37.6 | 1212 | 15.5 | ND | ND |
| Versus mouse ESTsc | 3244 | 37.9 | 1365 | 16.0 | ND | ND | 1787 | 22.9 | 509 | 6.5 | ND | ND |
aThe numbers were obtained from the results of an homology search against ESTs in the January 13, 2001 release of the GenBank database in which B(b)ovine, Bos taurus, B.taurus or cattle are contained in the entry name.
bThe homology search was performed against human ESTs in the January 13, 2001 release of the GenBank database.
cThe homology search was performed against mouse ESTs in the January 13, 2001 release of the GenBank database.
We described a new system to produce large numbers of high quality ESTs, with which we determined 600 ESTs per day. We obtained both 5′ and 3′ cDNA sequences derived from ∼6900 genes that will be used for RH mapping. The draft sequences of the human genome will facilitate finding human orthologs and determining their chromosomal positions (3,4), which will promote construction of a precise comparative map. This approach is applicable to other organisms whose ESTs need to be developed.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Toshio Watanabe for discussions and Hikaru Yamamoto and Ryohta Etoh (Hitachi Software Engineering Co. Ltd) for technical assistance. This study was supported by grants from the Japan Racing and Livestock Promotion Foundation.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +81 248 25 5641; Fax: +81 248 25 5725; Email: kazusugi@siag.or.jp AV588548–AV618892, AV662325–AV668289
REFERENCES
- 1.Lyons L.A., Laughlin,T.F., Copeland,N.G., Jenkins,N.A., Womack,J.E. and O’Brien,S.J. (1997) Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nature Genet., 15, 47–56. [DOI] [PubMed] [Google Scholar]
- 2.Deloukas P., Schuler,G.D., Gyapay,G., Beasley,E.M., Soderlund,C., Rodriguez-Tomé,P., Hui,L., Matise,T.C., McKusick,K.B., Beckmann,J.S. et al. (1998) A physical map of 30,000 human genes. Science, 282, 744–746. [DOI] [PubMed] [Google Scholar]
- 3. International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 4.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 5.Grobet L., Martin,L.J.R., Poncelet,D., Pirottin,D., Brouwers,B., Riquet,J., Schoeberlein,A., Dunner,S., Menissier,F., Massabanda,J. et al. (1997) A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nature Genet., 17, 71–74. [DOI] [PubMed] [Google Scholar]
- 6.Jeon J.-T., Carlborg,Ö., Törnsten,A., Giuffra,E., Amarger,V., Chardon,P., Andersson-Eklund,L., Andersson,K., Hansson,I., Lundström,K. and Andersson,L. (1999) A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. Nature Genet., 21, 157–158. [DOI] [PubMed] [Google Scholar]
- 7.Nezer C., Moreau,L., Brouwers,B., Coppieters,W., Detilleux,J., Hanset,R., Karim,L., Kvasz,A., Leroy,P. and Georges,M. (1999) An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nature Genet., 21, 155–156. [DOI] [PubMed] [Google Scholar]
- 8.Andersson L., Haley,C.S., Ellegren,H., Knott,S.A., Johansson,M., Andersson,K., Andersson-Eklund,L., Edfors-Lijia,I., Fredholm,M., Hansson,I. et al. (1994) Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science, 263, 1771–1774. [DOI] [PubMed] [Google Scholar]
- 9.Georges M., Nielsen,D., Mackinnon,M., Mishra,A., Okimoto,R., Pasquino,A.T., Sargeant,L.S., Sorensen,A., Steele,M.R., Zhao,X., Womack,J.E. and Hoeschele,I. (1995) Mapping quantitative trait loci controlling milk production in dairy cattle by exploiting progeny testing. Genetics, 139, 907–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coppieters W., Riquet,J., Arranz,J.-J., Berzi,P., Cambisano,N., Grisart,B., Karim,L., Marcq,F., Moreau,L., Nezer,C. et al. (1998) A QTL with major effect on milk yield and composition maps to bovine chromosome 14. Mamm. Genome, 9, 540–544. [DOI] [PubMed] [Google Scholar]
- 11.Kang’a S., Nilsson,P.H., Rottengatter,K., Goldammer,T., Kim,C.D., Srinivas,K., Iraqi,F., Mwakaya,J., Mwangi,D., Schwerin,M. et al. (2000) Comparative mapping of a cattle trypanotolerance QTL region on Bta7. In 27th International Conference on Animal Genetics Abstract Book. Blackwell Science Ltd, UK, p. 7.
- 12.Band M.R., Larson,J.H., Rebeiz,M., Green,C.A., Heyen,D.W., Donovan,J., Windish,R., Steining,C., Mahyuddin,P., Womack,J.E. and Lewin,H.A. (2000) An ordered comparative map of the cattle and human genomes. Genome Res., 10, 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawamoto S., Yoshii,J., Mizuno,K., Ito,K., Miyamoto,Y., Ohnishi,T., Matoba,R., Hori,N., Matsumoto,Y., Okumura,T. et al. (2000) BodyMap: a collection of 3′ESTs for analysis of human gene expression information. Genome Res., 10, 1817–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith T.P.L., Grosse,W.M., Freking,B.A., Roberts,A.J., Stone,R.T., Casas,E., Wray,J.E., White,J., Cho,J., Fahrenkrug,S.C. et al. (2001) Sequence evaluation of four pooled-tissue normalized bovine cDNA libraries and construction of a gene index for cattle. Genome Res., 11, 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aso H., Abe,H., Nakajima,I., Ozutsumi,K., Yamaguchi,T., Takamori,Y., Kodama,A., Hoshino,F.B. and Takano,S. (1995) A preadipocyte clonal line from bovine intramuscular adipose tissue: nonexpression of GLUT-4 protein during adipocyte differentiation. Biochem. Biophys. Res. Commun., 213, 369–375. [DOI] [PubMed] [Google Scholar]
- 16.Ewing B., Hillier,L., Wendl,M.C. and Green,P. (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res., 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 17.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 18.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozen S. and Skaletsky,H.J. (1998) Primer3. Code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html.
- 20. Comparative Genome Organization (1996) Comparative genome organization of vertebrates. Mamm. Genome, 7, 717–734. [DOI] [PubMed] [Google Scholar]