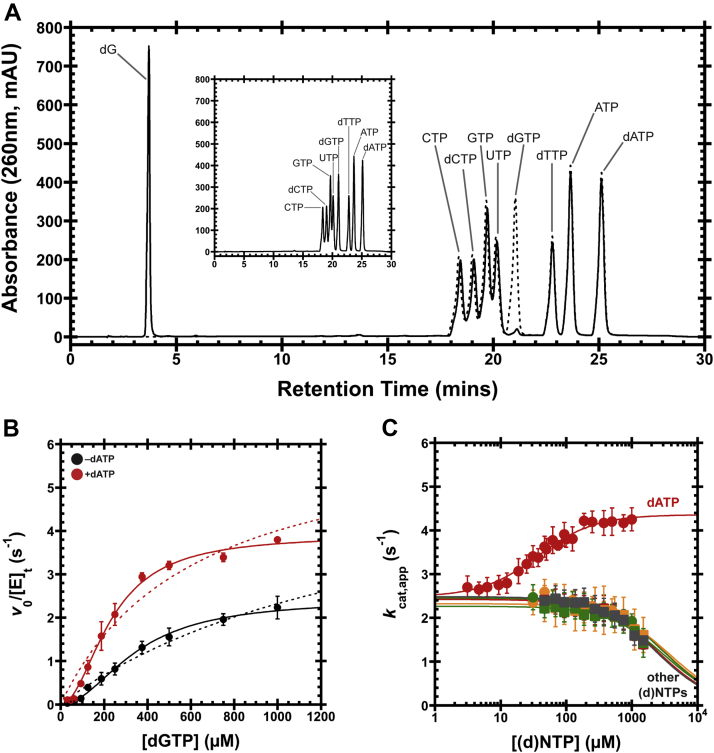
Figure 2.
Leeuwenhoekiella blandensis dGTPase activity and allosteric activation.A, 1-h hydrolysis of a mixture containing all eight standard dNTPs and NTPs by the dNTPase homolog from L. blandensis. The eight (d)NTPs were reacted together (1 mM each), with the substrates and products separated by HPLC (solid line). A control reaction at time 0 is shown as a dashed line and separately in the inset, for clarity. B, v0/[E]tversus [dGTP] plots with or without 500 μM dATP. Data are presented as the mean and SEM of three independent experiments. The solid lines are the fit of Equation 1 to the data with nH = 2, whereas the dashed lines are the fit for nH = 1. For dGTP-only, kcat = 2.6 ± 0.1 s−1, KM = 390 ± 30 μM, and nH = 1.7 ± 0.1. With 500 μM dATP, kcat = 3.8 ± 0.1 s−1, KM = 224 ± 9 μM, and nH = 2.2 ± 0.1. C, dGTPase activity at 1 mM dGTP, titrating the remaining seven standard (d)NTPs. We refer to this substrate concentration as an apparent kcat (kcat,app) because it is above the threshold required to bring the turnover within 20% of the kcat, thereby approximating conditions with saturating substrate. Data are plotted as the mean and SEM of four independent experiments. Activation was observed for dATP (red circles), with K1/2 = 36 ± 3 μM and nH = 1.3 ± 0.1. Inhibitory effects are observed for dTTP (orange circles), dCTP (green circles), ATP (red squares), UTP (orange squares), CTP (green squares), and GTP (gray squares). The IC50s are estimated to be >2 mM. dNTP, deoxynucleoside triphosphate; dNTPase, dNTP triphosphohydrolase.