Abstract
GuiLingJi (GLJ), a classic traditional Chinese medicine (TCM) formula, is composed of over 20 herbs, according to the Pharmacopeia of the People's Republic of China. Owing to its various activities, GLJ has been used in clinical settings for more than 400 years in China. However, the ambiguous chemical material basis limits the development of studies on the quality control and pharmacological mechanisms of GLJ. Therefore, comprehensive characterization of the multiple chemical components of GLJ is of great significance for the modernization of this formula. Given the great variety of herbs in GLJ, both UHPLC-MS and 1H NMR techniques were employed in this study. In addition, solvent extraction with different polarities was used to eliminate signal interference and the concentration of trace components. A variety of MS analytic methods were also used, including implementation of a self-built compound database, diagnostic ion filtering, mass defect filtering, and Compound Discoverer 3.0 analysis software. Based on the above strategies, a total of 150 compounds were identified, including 5 amino acids, 13 phenolic acids and glycosides, 11 coumarins, 72 flavones, 20 triterpenoid and triterpenoid saponins, 23 fatty acids, and 6 other compounds. Moreover, 13 compounds were identified by 1H NMR spectroscopy. The UHPLC-MS and 1H NMR results supported and complemented each other. This strategy provides a rapid approach to analyzing and identifying the chemical composition of Chinese herbal prescriptions. The current study provides basis for further research on the quality control and pharmacological mechanism of GLJ.
Keywords: GuiLingJi, Chemical component, UHPLC-MS, 1H NMR, Traditional Chinese medicine
Graphical abstract
Highlights
-
●
The integrated approach of UHPLC-MS and 1H NMR techniques coupled with polarity partition strategy has been used for comprehensively characterizing the multiple chemical components of GLJ.
-
●
A variety of HRMS analytic methods used included self-built compounds database, diagnostic ions filtering, mass defect filtering, and software analysis for rapid identification the chemical components of GLJ.
-
●
The 163 compounds including flavones, phenolic acids and glycosides, triterpenoid and triterpenoid saponins, coumarin, fatty acids, amino acids, organic acids, organic bases and sugars were rapidly identified, and to clarify the chemical material basis of GLJ.
-
●
Established an analysis strategy which could be applied to other TCM formula for comprehensive characterization and identification of chemical components.
1. Introduction
Traditional Chinese medicine (TCM) formulas have been used in clinical settings for thousands of years in China. However, although the clinical efficacy of TCM formulas is definite, their ambiguous effective substances pose a main obstacle to the modernization of these formulas [1]. TCM formulas apply a multicomponent and multitarget approach in the treatment of diseases, which is different from Western medicines, and the pharmacologic mechanisms have been increasingly clarified. Therefore, comprehensive characterization and identification of the multiple components of TCM formulas is of particular importance.
Recently, UHPLC-MS has been increasingly used for rapid chemical profiling of complicated medicinal herbs. This technique is especially suitable for the identification of unknown organic compounds because of its high resolution, high sensitivity, high selectivity, and accurate mass values [[2], [3], [4]]. With the MS/MS technique, accurate mass values can help elucidate the molecular weight and candidate chemical formula. Moreover, both elemental composition and fragment ions can be obtained, which significantly reduces the number of possible structures of putative compounds and enables the efficiency analysis of complex samples. The diagnostic ion and mass defect filtering strategy minimizes ubiquitous matrix interference, extracts the MS features of some potentially active compounds, and rapidly identifies a series of compounds with the same skeleton [5]. However, MS-based approaches can only identify the components by comparison with those of reference compounds, so they are only suitable for molecules that are easily ionized. NMR spectroscopy is one of the most definitive identification methods and shows non-selectivity. However, the lower sensitivity and the overlaps of signals in the complex matrix limitits applications in identifying the chemical components of TCM formulas. Combining NMR and UHPLC-MS methods to identify the matrix chemical components of TCM formulas can facilitate the final characterization [6,7] and provide a new strategy for analyzing the complex systems of TCM formulas.
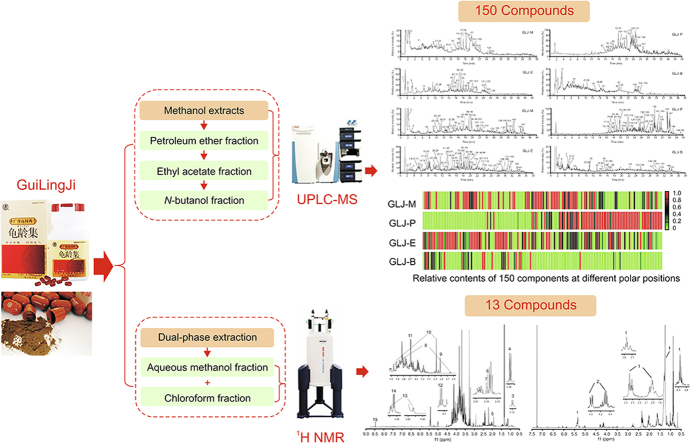
GuiLingJi (GLJ) is a well-known and classical TCM formula that can be traced back hundreds of years ago to the Ming and Qing dynasties, where it was used as an elixir. Modern pharmacological studies on GLJ indicate that it can significantly improve kidney-yang deficiency syndrome and testicular dysfunction [8,9], promote spermatogenesis in oligospermia rats by increasing the density and amount of sperms [10], and improve the mitochondrial function of sperms in idiopathic asthenospermia and the level of sperm viability [11]. Additionally, GLJ can improve the impairment of learning and memory functions caused by aging [12,13]. GLJ has various medical efficacies based on its multiple components. According to the Chinese Pharmacopeia (2020 edition), GLJ is one of the confidential products of national brands of TCM with 20 opened herbs, including Ginsen Radix et Rhizoma Rubar, Cervi Cornu Pantotrichum, Hippocampus, Lycii Fructus, Caryophylli Flos, Cyathulae Radix, Cynomorii Herba, Rehmanniae Radix Praeparata, Psoraleae Fructus, Cuscutae Semen, Eucommiae Cortex, Cistanches Herba, Glycyrrhizae Radix Et Rhizoma, Asparagi Radix, Epimrdii Folium, and Amomi Fructus, among others [14]. Each herb has its own unique processing method. For instance, Cervi Cornu Pantotrichum is fried with mature vinegar, Lycii Fructus is fried with honey, and Cistanches Herba is fried with yellow wine. More than 20 kinds of single herbs are processed and mixed with powder. Then, different from traditional decocting or extraction methods, GLJ is made of the above mixed powder by means of alchemy. For such an exclusive and complicated TCM formula possessing precise curative effects, few studies have investigated the systematic characterization of chemical components in GLJ. Therefore, clarifying the material basis of GLJ will provide useful chemical information for quality control and mechanism-of-action research.
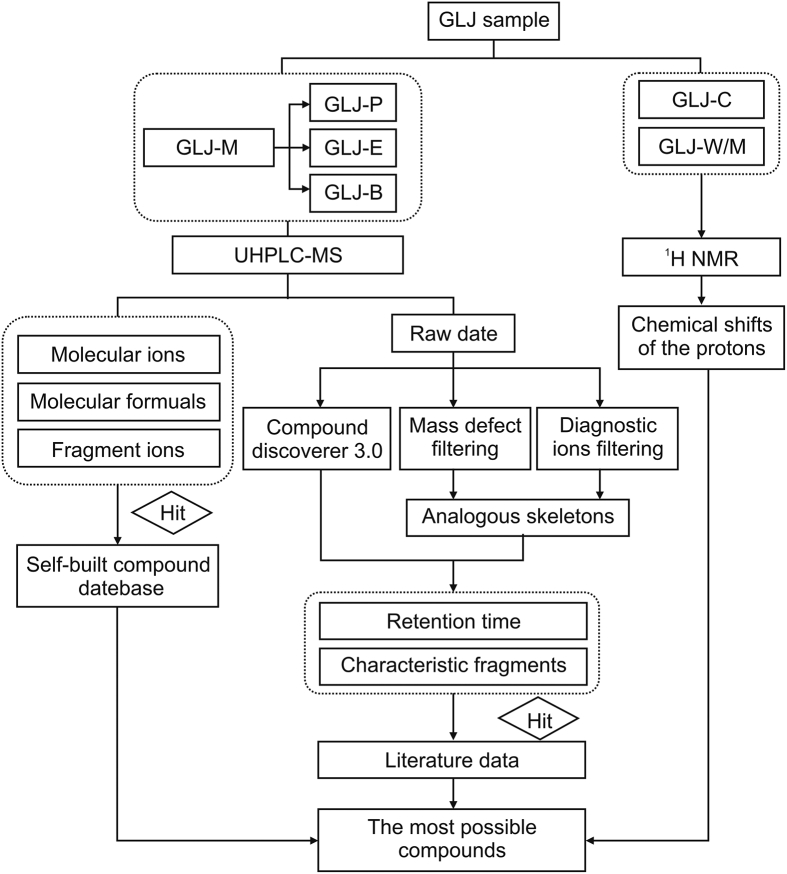
In this study, an enhanced identification strategy was developed for comprehensive characterization of the multiple chemical components of GLJ. First, the chemical components of the 16 herbs in the prescription were summarized to establish a self-built database of internal compounds to predict the chemical components of GLJ. Second, the methanol extract of GLJ was partitioned into different polar parts and characterized with UHPLC-MS techniques. A series of methods, such as summarizing the fragmentation regularity, diagnostic ion filtering, and mass defect filtering and inference by Compound Discoverer 3.0 (CD) software, was used to analyze the mass spectrometric data. Moreover, chemical compounds of GLJ were identified by comparing the retention time and MS/MSn data with those of individual herbs. Furthermore, two polar fractions of GLJ obtained by two-phase extraction were characterized by 1H NMR spectroscopy. Finally, a series of methods was combined to explore the complicated chemical composition of GLJ. The framework of this study is displayed in Fig. 1.
Fig. 1.
Summary diagram of the combined strategy of UHPLC-MS and 1H NMR analysis. GLJ: GuiLingJi; GLJ-M: the methanol extract of GLJ; GLJ-P: the petroleum ether fraction; GLJ-E: ethyl acetate fraction; GLJ-B: the n-butanol fraction; GLJ-C: the chloroform fraction; GLJ-W/M: the aqueous methanol fraction.
2. Experimental
2.1. Reagents and materials
GLJ (batch No.: 20170726) was generously provided by Shanxi Guangyuyuan Traditional Chinese Medicine Co., Ltd. (Jinzhong, China). Standard references included kaempferol (≥98%, batch No.: MUST-17032911), psoralen (≥98%, batch No.: MUST-16092810), baohuoside Ⅰ (≥98%, batch No.: MUST-19042203), isobavachalcone (≥98%, batch No.: MUST-16102615), neobavaisoflavone (≥98%, batch No.: MUST-16092810), bavachinin A (≥98%, batch No.: MUST-18101705), oleanolic acid (≥98%, batch No.: MUST-17042706), ginsenoside Re (≥98%, batch No.: 180829), and ginsenoside Rg1 (≥98%, batch No.: 180917), which were obtained from Chengdu Must Biotechnology Co., Ltd. (Chengdu, China) and Shanghai Ronghe Pharmaceutical Technology Development Co., Ltd. (Shanghai, China). Methanol, petroleum ether, ethyl acetate, and n-butanol of analytical grade were purchased from Tianjin Four Fine Chemicals Co., Ltd. (Tianjin, China). Ultrapure water was prepared with a Milli-Q water purification system (Millipore, Bedford, MA, USA). Methanol of LC-MS grade and formic acid of HPLC grade were obtained from Thermo Fisher Scientific Inc. (Thermo Fisher Scientific, Waltham, MA, USA). Deuterated chloroform (CDCl3, 99.8% D) containing tetramethylsilane (TMS, 0.03%, m/V) and methanol-d4 (99.8% D) were obtained from J&K Scientific Ltd. (Beijing, China).
2.2. Sample preparation
The reference standards (0.1–1.0 mg/mL) were accurately weighed and then mixed and dissolved in methanol of LC-MS grade. An accurately weighed 1.00 g sample of GLJ powder was ultrasonically extracted with 50 mL of methanol at room temperature for 30 min. The extracts were filtered and concentrated to be dried. One part of the residue was transferred to a 5 mL volumetric flask and dissolved in LC-MS grade methanol to volume as the methanol extract of GLJ (GLJ-M). The 16 herbs were extracted under the same process as GLJ-M to identify the source of ingredients. Meanwhile, the other part of the residue was suspended in 30 mL of water and successively treated with petroleum ether, ethyl acetate, and n-butanol. Then, the petroleum ether fraction (GLJ-P), ethyl acetate fraction (GLJ-E) and n-butanol fraction (GLJ-B) were concentrated to dryness. The residue was dissolved in LC-MS grade methanol, and the solution was completely transferred into a 2 mL volumetric flask. All solutions were filtered through a nylon syringe filter (0.22 μm) for UHPLC-MS analysis.
For 1H NMR analysis, 1.00 g of GLJ powder was exactly weighed and ultrasonically extracted with 24 mL of extraction solvent composed of water, methanol, and chloroform at a volume ratio of 1:1:2 at room temperature for 30 min. The extracts were centrifuged at 3,500 r/min for 20 min. Then, the two layers were separated and concentrated to dryness, and the residues were obtained. The chloroform fraction (GLJ-C) was dissolved in 800 μL of CDCl3, and the aqueous methanol fraction (GLJ-W/M) was dissolved in 800 μL of CD3OD. Then, the resulting mixture was centrifuged for 15 min at 13,000 r/min. Finally, 600 μL of supernatant was transferred to an oven-dried 5 mm NMR tube for 1H NMR analysis.
2.3. UHPLC-MS conditions
UHPLC-MS analysis was conducted on a Thermo-Fisher UHPLC system coupled to a Q Exactive Orbitrap high-resolution mass spectrometer (UHPLC-QE HRMS; Thermo Fisher Scientific, Waltham, MA, USA). Chromatographic separation was carried out on a Waters Acquity UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 μm; part No.: 186003539) maintained at 30 °C. The mobile phase of solvent A (water with 0.1% formic acid) and solvent B (acetonitrile) was operated under the following program: 0–10 min, 5%–30% B; 10–15 min, 30%–52% B; 15–18 min, 52% B; 18–26 min, 52%–87% B, 26–30 min, 87% B; 30–35 min, 87%–100% B; and 35–38 min, 100%–5% B. The sample injection volume was 3 μL, and the flow rate was 0.2 mL/min.
The mass spectrometer was fitted with an electrospray ionization source, and mass detection was carried out in both positive and negative ion modes with the following settings: capillary temperature, 320 °C; heater temperature, 300 °C; sheath gas velocity, 35 arb; auxiliary gas flow rate, 10 arb; full scan range, m/z 100–1500; and MS2 scan range, m/z 50–1500. Fast data-dependent acquisition was applied to trigger MS/MS acquisition of precursor ions with an intensity threshold of 5000. Full scan spectra were acquired at a scan time of 0.3 s, followed by MS/MS spectra acquisition at a scan time of 0.1 s. The top 15 most intense ions were selected to perform MS/MS acquisition. The collision energy was set to a ramp of 25–60 V for MS/MS acquisition.
2.4. 1H NMR conditions
1H NMR spectra were measured at 600.13 MHz using an Avance 600 Bruker spectrometer (Bruker Biospin, Rheinstetten, Germany) equipped with a Bruker 5 mm double resonance BBI probe. CD3OD and CDCl3 were used for internal lock purposes. Spectra of GLJ-C and GLJ-W/M were acquired using the zg30 sequence. Sixty-four scans were recorded, resulting in a total acquisition time per sample of 5 min.
2.5. Data processing
The UHPLC-QE HRMS raw data were automatically processed using an Xcalibur workstation (Thermo Fisher Scientific, Waltham, MA, USA). Then, the processed data were imported into CD software (Thermo Fisher Scientific, Waltham, MA, USA) for peak extraction and normalization. The peak width at 5% height and the peak-to-peak baseline noise were automatically determined, and the processing parameters were set as follows: retention time range, 0–38 min; mass range, 100–1500 Da; mass tolerance, 5 ppm; retention time tolerance, 0.05 min; S/N threshold, 1.5; negative adducts, [M−H]−, [M + HCOO]−; and positive adducts, [M+H]+ and [M+Na]+. The compounds characterized by CD software were identified by searching the mz Cloud, Masslist, Chemspider, and PubChem databases.
The NMR spectra were processed using MestReNova (version 6.1.1, Mestrelab Research, Santiago de Compostella, Spain) to manually correct the baseline and phase and calibrated to TMS at δ 0.00 for CDCl3. All spectra were referenced to the residual signal of CD3OD at δ 3.31.
3. Results and discussion
3.1. Establishment of compound databases and analysis of fragmentation rules
According to Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (https://tcmsp-e.com/tcmsp.php), Web of Science (https://www.isiknowledge.com/), and CNKI (http://www.cnki.net/), a database of compounds containing compound names, molecular formulas, accurate molecular masses, and chemical structures was established. The established compound database included 16 of the medicinal herbs in GLJ without some mineral and animal drugs, as the chemical compositions of these mineral and animal drugs had not been studied. In addition, mass spectrum fragment ions of some compounds were obtained by Massbank (http://massbank.eu/) and the literature, and the 1H NMR chemical shifts of some primary metabolites were obtained by HMBD (https://hmdb.ca/). Finally, a total of 1246 compounds including 358 flavonoids, 179 triterpenes and their glycosides, 143 phenylpropanes and their glycosides, 123 steroids and their glycosides, 98 organic acid, 54 saccharides, 20 amino acids, and 271 other compounds (alkaloids, tannins, polyphenols, unsaturated hydrocarbons, etc.) were summarized.
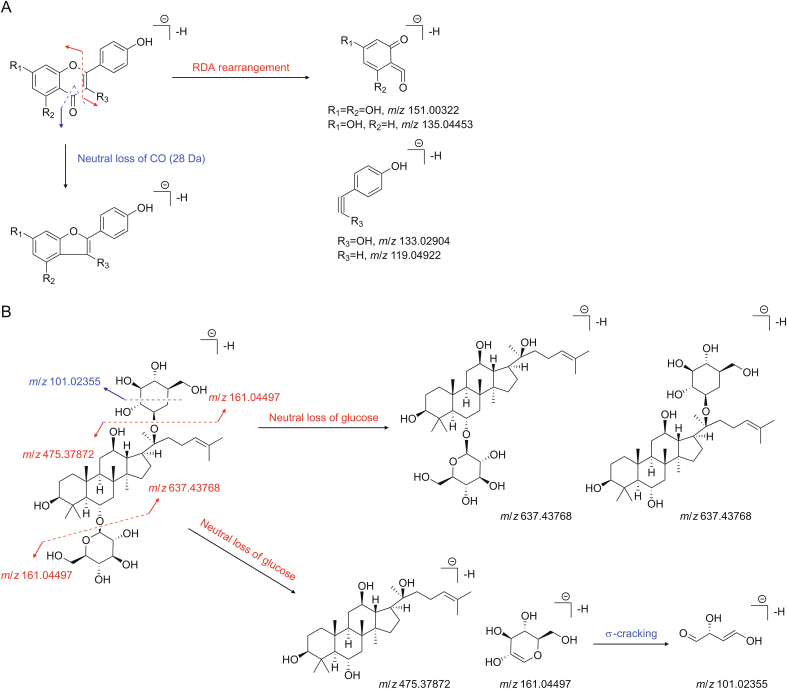
The fragmentation rules of the compounds were also studied, taking flavonoids and ginsenoside as examples. Representative authentic standards of flavonoids, including kaempferol, baohuoside Ⅰ, isobavachalcone, neobavaisoflavone and bavachinin A, were subjected to analysis by a UHPLC-QE HRMS system in both positive and negative ion modes. Fragmentation patterns of each standard were summarized based on assigned fragment ions. The prominent product ions were selected as diagnostic ions. The prime diagnostic ions or neutral loss fragments of these compounds were confirmed by MS/MS spectra analysis of 7 standards as well as by the self-built compound database. Because the characteristic fragmentation pattern of flavonoids indicated a retro-Diels-Alder (RDA) reaction, significant product ions of the RDA reaction at m/z 119.04922, 135.04453, 133.02904, and 151.00322 were set as diagnostic ions. The possible fragment pathways of flavonoids in negative ion mode are shown in Fig. 2A.
Fig. 2.
Possible fragment pathways and diagnostic ions of (A) flavonoids and (B) ginsenoside.
From the fragment ions of the reference standard including ginsenosides Re and Rg1 and the reported literature [[15], [16], [17]], fragmentation rules could be found. The protopanaxadiol, protopanaxatriol, and oleanolic acid types of ginsenosides were found at m/z 459, 475, and 455, corresponding to successive losses of sugar moieties, respectively. Characteristic neutral losses of these sugar residues were 162 Da (hexose), 146 Da (deoxyhexose), and 132 Da (fructose). The lost sugar units, e.g., glucose, rhamnose, and xylose, produced common highly abundant ions at m/z 101, 113, 119, 161, and 179. Based on the above fragmentation regularity, prominent product ions such as m/z 455, 459, and 475 were selected as diagnostic ions of the analogous skeletons, and m/z 101, 113, 119, 161, and 179 were selected as diagnostic ions of the sugar residues. The possible fragment pathways of ginsenosides in negative ion mode are shown in Fig. 2B, with ginsenoside Rg1 as an example.
3.2. UHPLC-QE HRMS analysis and identification of the main constituents in GLJ
Prior to real sample analysis, extraction methods and chromatography conditions were fully evaluated and optimized. To achieve better chromatographic separation and mass spectrometric detection, different extraction solvents including 100% methanol, 75% methanol, and 100% water were explored. The extracts with 100% methanol showed higher peak responses but poor peak separations; thus, solvents with different polarities were used. Various compositions of mobile phases such as methanol-water, acetonitrile-water, and acetonitrile-water with formic acid were explored. Acetonitrile-water with formic acid was chosen as the mobile phase due to its more symmetrical peak shape and ionization efficacy in both positive and negative ion modes.
Methodological verification was carried out to monitor the stability of the LC-MS system. Ten ions with different retention time and m/z values from the base peak intensity chromatograph of 6 GLJ-M samples were extracted. The RSDs for retention time were 4.88 × 10−4%–3.77 × 10−3%, the m/z values were 3.64 × 10−6%–4.85 × 10−5% and the relative peak areas were 5.02%–12.94% (Table S1), which demonstrated the favorable stability of the system and the reliability of the data analysis.
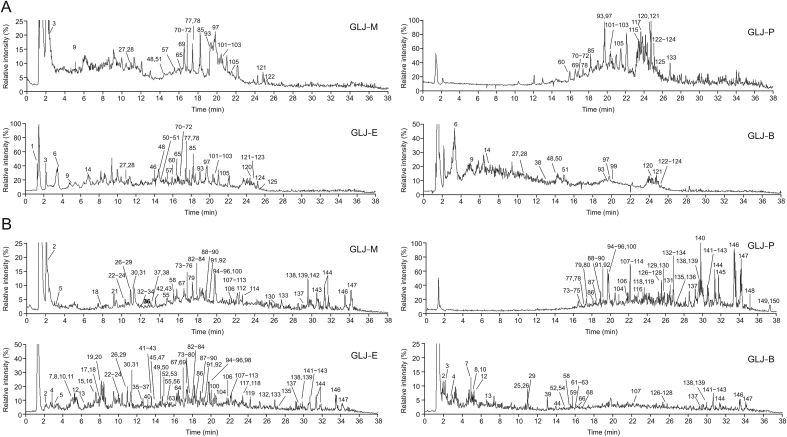
A total of 150 compounds were tentatively characterized, including 5 amino acids, 13 phenolic acids and glycosides, 11 coumarin, 72 flavone, 20 triterpenoid and triterpenoid saponins, 23 fatty acids and 6 others. Among them, 9 components were definitively identified by comparing their reference standards, while the others were tentatively identified by comparing the accurate masses, ESI-MSn fragment ions, and molecular weights with those in the self-built compound database. CD software provided some references including matched data and fitting compounds. The total ion chromatograms of GLJ-M, GLJ-P, GLJ-E, and GLJ-B in both negative and positive ion modes are shown in Fig. 3. All the compounds that we identified in GLJ were already known; no new compounds were identified. The corresponding chromatographic and mass spectral data of 150 compounds are shown in Tables S2–S13. The source of ingredients was identified by comparing the accurate masses and retention time with the 16 herbs. Some amino acids and fatty acids may be mainly derived from multiflavor animal medicines without exclusive sources. The exact peaks with mass ranges and ion fragments of standard references and GLJ are shown in (Figs. S1–S9).
Fig. 3.
The total ion chromatograms of GLJ-M, GLJ-P, GLJ-E, GLJ-B in (A) the positive and (B) the negative ion modes.
3.2.1. Identification of amino acids in GLJ
Five amino acids were identified in GLJ (Table S2). Amino acids might lose COOH in the reaction of α-cracking. For instance, compound 3 was detected at 2.42 min, which indicated that it is a polar compound. The molecular formula of compound 3 was determined to be C9H11NO3 according to the result that the typical MS/MS spectra of the protonated ions at m/z 182.08119 [M+H]+ produced the ions at m/z 136.07578 and 119.04925 due to successive losses of COOH and NH2 (Fig. 4). Compound 3 was identified as tyrosine by comparing the fragment ions with those in the literature [18]. Other amino acids were inferred by CD and further identified by comparing the MS/MSn data with those in the literature [18]. The chemical structures of amino acids in GLJ are shown in Fig. S10.
Fig. 4.
The MS/MS spectra and proposed fragmentation for tyrosine in positive ion mode.
3.2.2. Identification of phenolic acids and glycosides in GLJ
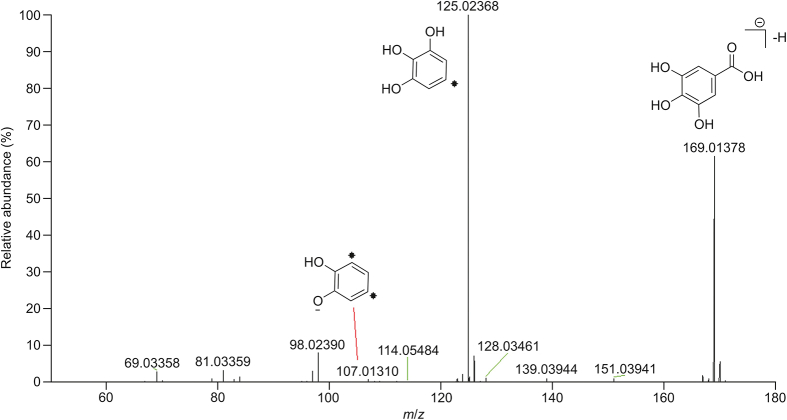
Thirteen phenolic acids and glycosides were identified in GLJ in negative ion mode (Table S3). The characteristic fragmentation pattern of phenolic acids was neutral losses of CO2, OH or COOH. For instance, compound 2 was deduced as gallic acid by CD software. Its retention time was 2.10 min, and the molecular formula was determined to be C7H6O5. The typical MS/MS spectra of the deprotonated ions at m/z 169.01378 [M−H]− produced ions at m/z 125.02368 [M−H–CO2]− and 107.01310 [M−H–CO2–OH]−. The MS/MS2 spectra and fragmentation patterns are shown in Fig. 5. Compound 2 was identified as gallic acid by comparing the fragment ions with those in the literature [6]. Other phenolic acids were identified by comparing the exact mass, molecular formulas and fragment ions with those in a self-built compound database.
Fig. 5.
The MS/MS spectra and proposed fragmentation for gallic acid in negative ion mode.
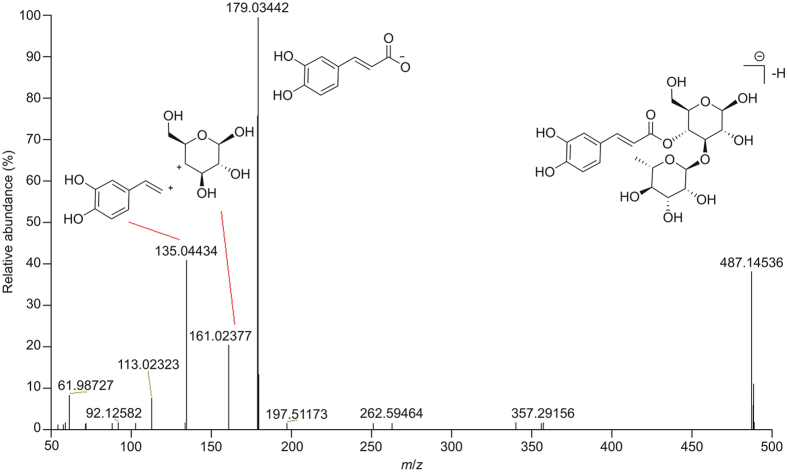
Five phenylpropanoid glycosides were identified in GLJ. For example, compound 13 displayed an [M−H]− ion at m/z 487.14587 and generated fragment ions at m/z 179.03442 and 161.02377. The fragment ion at m/z 179.03442 was a caffeoyl species and was generated by neutral losses of 162 Da (hexose) and 146 Da (deoxyhexose). The fragment ions at m/z 161.02377 confirmed the existence of hexose (Fig. 6). Accordingly, compound 13 was identified as a phenylpropanoid glycoside named cistanoside F. Then, compounds 21, 23, 29, and 45 were filtered by taking the fragment ions m/z 179 as diagnostic ions or the mass defect of 162,146 Da. These compounds were identified as echinacoside, caffeic acid 3-glucoside, acteoside, and ferulic acid-4-β-glucoside by comparing the fragment ions and molecular formulas with the literature [19]. The chemical structures of phenolic acids and glycosides in GLJ are shown in Fig. S11.
Fig. 6.
The MS/MS spectra and proposed fragmentation for cistanoside F in negative ion mode.
3.2.3. Identification of coumarins in GLJ
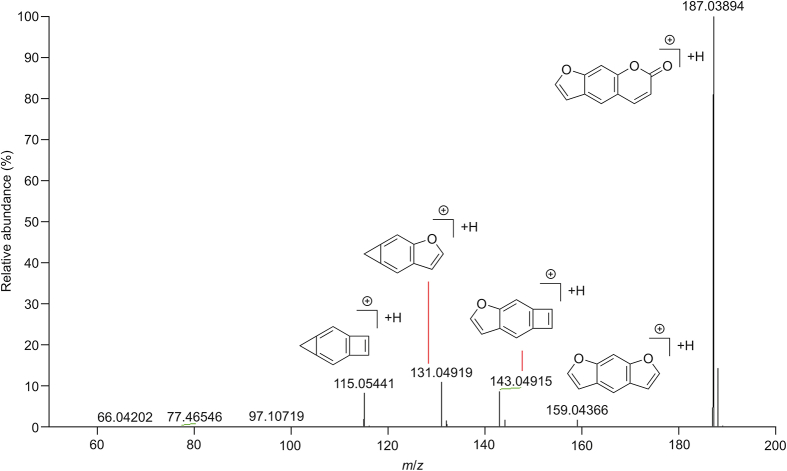
Eleven coumarins were identified in GLJ (Table S4). The main MS/MS behavior of coumarins was neutral loss of 28 Da (CO) and 44 Da (CO2). For example, compound 72 was identified by comparing the retention time and ion fragments with the reference compound. Protonated ions at m/z 187.03894 [M+H]+ were observed, which further generated the ions at m/z 159.04366 [M+H–CO]−, m/z 143.04915 [M−H–CO2]−, m/z 131.04919 [M−H–CO–CO]– and 115.05441 [M+H–CO2–CO]−. The mass spectra and proposed major fragmentation with structures are shown in Fig. 7. Similarly, compound 90 displayed a fragment ion at m/z 367.11911 [M−H]−. Then, it generated ions at m/z 352.09702 [M−H–CH3]− and m/z 297.04062 [M−H–C5H10]−, which provided methyl and isopentane groups. The characteristic fragment ions at m/z 324.10007 [M−H–CH3–CO]− and m/z 269.04532 [M−H–C5H10–CO]− were explained by the dissociation of the lactone ring. Compound 90 was identified as isoglycycoumarin based on the above MS/MSn data. Thus, compounds 43, 56, 74, 81, 87, 89, 106, 110, and 119 were tentatively identified by comparing the MS/MSn data with those in the literature [[20], [21], [22]]. The chemical structures of coumarins in GLJ are shown in Fig. S12.
Fig. 7.
The MS/MS spectra and proposed fragmentation for psoralen in positive ion mode.
3.2.4. Identification of flavones in GLJ
A total of 72 compounds were characterized as flavonoids, including flavonols, flavones, chalcones, isoflavones, flavanones, and flavone glycosides. These flavonoids with different structural skeletons had different characteristic fragment ions.
3.2.4.1. Identification of flavonols
Eleven flavonols were identified in GLJ (Table S5). Compound 71 was determined to be kaempferol by comparing the retention time and ion fragments with the reference compound. Compound 71 displayed deprotonated ions at 285.04077 [M−H]− and produced ions at m/z 257.04501 by the neutral loss of CO. A series of ions at m/z 151.00322 and m/z 133.02904 were generated by RDA cleavage. Similarly, the mass spectra of compound 5 showed an abundant [M−H]− ion at m/z 301.03488, which generated a product ion at m/z 273.04028 by neutral loss of CO, a product ion at m/z 178.99866 by loss of the B-ring and CH, and a product ion at m/z 151.00308 by the RDA reaction. Compound 5 was identified as quercetin by comparing the fragment ions with those in the literature [23]. Other flavonols were filtered by taking the fragment ions at m/z 285, 255, 151, 301, and 178 as diagnostic ions. Then, compounds 10, 16, 52, 60, 73, 94, 95, 96, and 104 were further identified by comparing the MS/MSn data with the literature [23,24]. The chemical structures of flavonols identified in GLJ are shown in Fig. S13.
3.2.4.2. Identification of flavones
Ten flavones were identified in GLJ (Table S6). For example, the retention time of compound 55 was 14.99 min, and its molecular formula was C15H10O6. Compound 55 displayed a [M−H]− ion at m/z 285.04077. In addition, the fragment ions at m/z 257.04565 and m/z 241.35442 were generated by successive loss of CO and O, respectively, and the fragment ions at m/z 151.00298 and m/z 133.02856 were generated by RDA cleavage. The relative abundance of m/z 133.02856 was higher than that of m/z 151.00298, which is different from the fragment ion abundance of kaempferol. Compound 55 was identified as luteolin by comparing the MS/MSn data with those in the literature [25]. Compounds 69, 80, and 100 were filtered by taking the fragment ion m/z 151 as the diagnostic ion and further identified by comparing the MS/MSn data with those in the literature [23,26,27]. Other flavones were tentatively characterized by comparing the retention time and ion fragments with those in the database of internal compounds. The chemical structures of flavones in GLJ are shown in Fig. S14.
3.2.4.3. Identification of chalcone
Eight chalcones were identified in GLJ (Table S7). Compound 103 was determined to be isobavachalcone by comparing the retention time and ion fragments with those of the reference compound. Compound 103 displayed an [M−H]− ion at m/z 323.12918 and a fragment ion at m/z 268.07498 [M−H–C4H7]−, indicating the existence of an isopentane group. The fragment ions at m/z 203.07111 and m/z 119.04950 were generated from the α-broken keto groups. Compounds 93, 112 and 122 were filtered by taking the fragment ion m/z 119 as the diagnostic ion and further identified by comparing the MS/MSn data with the literature [21,28]. Other chalcones were tentatively characterized by comparing the exact mass and ion fragments with those in the self-built compound database. The chemical structures of chalcones in GLJ are shown in Fig. S15.
3.2.4.4. Characterization of isoflavones
Twenty isoflavones were identified in GLJ (Table S8). Compound 97 was determined to be neobavaisoflavone by comparing the retention time and ion fragments with those of the reference compound. Compound 97 displayed an [M+H]+ ion at m/z 323.12775 and a fragment ion at m/z 267.06509 [M + H–C4H7]−, indicating the existence of an isopentene group. The fragment ion at m/z 239.06970 [M + H–C4H7–CO]− was observed by neutral loss of CO successively, and the fragment ion at m/z 137.02341 was generated from RDA cleavage. Other isoflavones were filtered by taking the fragment ion m/z 151 as the diagnostic ion and further identified by comparing the MS/MSn data recorded in the literature [24,[29], [30], [31]]. The chemical structures of isoflavones contained in GLJ are shown in Fig. S16.
3.2.4.5. Characterization of flavanone
Seven flavanones were identified in GLJ (Table S9). Compound 121 was determined to be bavachinin A by comparing the retention time and ion fragments with those of the reference compound. Compound 121 has a precursor ion at m/z 339.15903 [M+H]+. The product fragment ions at m/z 271.09631 [M−C5H9]+ were generated by the loss of the isopentane group. The fragment ions at m/z 219.10146 [M + H–C8H8O]+, m/z 151.03885 [M + H–C5O9–C8H8O]+, and m/z 119.04922 [M + H–C13H14O3]− were the characteristic ions of RDA cleavage. The other 6 compounds were filtered by taking the fragment ions m/z 135 and 119 as diagnostic ions and identified by comparing the MS/MSn data in the literature [20,21,28]. The chemical structures of flavanone in GLJ are shown in Fig. S17.
3.2.4.6. Characterization of flavone glycosides
Sixteen flavone glycosides were identified in GLJ (Table S10). Flavone glycosides showed characteristic neutral losses of 176 Da (glucuronic acid, GlcA), 162 Da (glucosyl, Glu), 146 Da (rhamnosyl, Rha), and 132 Da (arabinosyl, Ara). The aglycon structure might lose CO, CO2, CHO and so on during RDA cleavage. For example, compound 91 was determined to be baohuoside Ⅰ by comparing the retention time and ion fragments with those of the reference compound. Compound 91 has a precursor ion at m/z 513.17688 [M−H]−. The fragment ion at m/z 366.11108 was generated by characteristic neutral losses of 146 Da (rhamnose); then, the fragment ions at m/z 351.08771 and m/z 323.09189 were generated by successive losses of CH3 and CO. Other flavone glycosides were filtered by using 176, 162, 146, and 132 Da as mass defects; these compounds were tentatively identified by comparing the MS/MSn data with those in the literature [[32], [33], [34], [35], [36], [37]]. The chemical structures of flavone glycosides in GLJ are shown in Fig. S18.
3.2.5. Identification of triterpenes and triterpenoid saponins in GLJ
Six triterpenes and 14 triterpenoid saponins were identified in GLJ (Table S11). Compound 142 was determined to be oleanolic acid by comparing the retention time and ion fragments with those of the reference compound. The deprotonated ion at m/z 455.35361 produced ions at m/z 407.32980 [M−H–CO2–4H]− and 391.25125 [M−H–CO2–2H–H2O]−. The fragment ion at m/z 247.28014 [M−H–C14H24O]− was the characteristic ion of RDA cleavage. Fragment ions such as [M−H–CO2]− and [M−H–HCOOH]− were regarded as the characteristic ions of triterpenes. Compounds 65, 116, 129, 130, and 132 were tentatively characterized as triterpene acids based on the above fragment rules [27,38].
The triterpenoid saponins in GLJ were mainly ginsenosides. Ginsenosides, such as ginsenoside Rg1, ginsenoside Rb, ginsenoside Re, and ginsenoside Rd, which have relatively higher contents in ginseng, were detected in the GLJ-M and GLJ-E fractions. However, many ginsenosides such as ginsenoside Ro, ginsenoside Rf, and ginsenoside Rc with relatively low contents could be detected only in the n-butanol fraction. As an example, compound 38 was determined to be ginsenoside Re by comparing the retention time and ion fragments with those of the reference compounds. Typical MS/MS spectra of the deprotonated ions of the standard reference samples at m/z 991.54944 [M−H + HCOOH]− were assigned to ginsenoside Re. The deprotonated ion at m/z 945.54315 produced ions at m/z 783.48767, 637.43170, and 475.37979 due to successive losses of glucose, rhamnose, and glucose. The structures of the other 12 ginsenosides were screened out by the diagnostic ion strategy and further identified by comparing the MS/MSn data with those in the literature [39,40]. The chemical structures of triterpenes and triterpenoid saponins in GLJ are shown in Fig. S19.
3.2.6. Identification of fatty acids in GLJ
Twenty-three fatty acids were identified in GLJ (Table S12). Fatty acids were mainly detected in the GLJ-P fraction. These compounds could be rapidly identified by characteristic ions including [M−H–CO]−, [M−H–CO2]−, [M−H–H2O]−, [M−H–COOH]−, [M−H–COOH]−, [M−H–CH]−, [M−H–CH2]−, and [M−H−CnH2n]−. Take compound 138 as an example. It had a retention time of 29.67 min, and the molecular formula was C18H30O2. Compound 138 had a precursor ion at m/z 277.21759 [M−H]− and generated product ions m/z 259.20721 and m/z 233.22884 by neutral loss of H2O and CO2, respectively. The fragment ions at m/z 84.41108, m/z 71.01276, and m/z 59.01290 were the characteristic ions of aliphatic and unsaturated aliphatic compounds. Compound 138 could be identified as α-linolenic acid or γ-linolenic acid by comparing the MS/MSn data with MassBank. Other fatty acids were inferred by CD software and then identified by comparing the MS/MSn data with MassBank data.
3.3. 1H NMR analysis of different polar parts of GLJ
To obtain a better separation degree and more compound information, various extraction conditions were investigated, including solvent extraction with different polarities and two-phase extraction. Eventually, the two-phase extraction with water/methanol and chloroform was selected because the 1H NMR spectra obtained from this extraction showed a higher peak response value and better separation degree and covered all distinct regions such as organic acids, amino acids, and phenolic regions.
A total of 13 compounds, namely, 6 organic acids or their ester, 2 amino acids, 2 organic bases, and 3 sugars, were characterized by 1H NMR spectroscopy. Moreover, 1H NMR analysis supported the existence of fatty acids and triglycerides.
3.3.1. Characterization of chloroform fraction
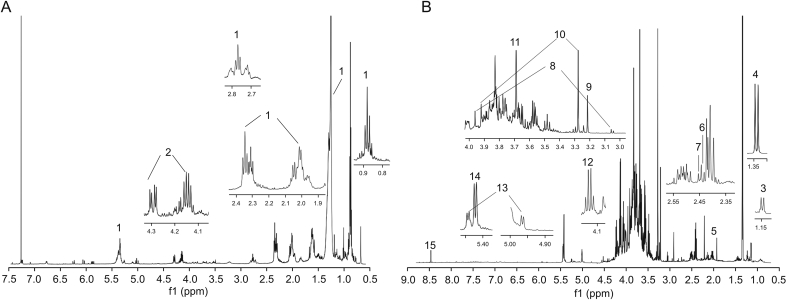
The 1H NMR spectrum of GLJ-C showed strong signals in the region from δ H 0.5 to δ H 3.2 ppm, where most signals were derived from amino acids and organic acids (Fig. 8A). The terminal methyl (δ 0.880, t, J = 7.2 Hz), methylene (δ 2.346, t, J = 7.2 Hz), allylic CH2 (δ 2.015, m), diallylic CH2 (δ 2.771 t, J = 6.6 Hz), other hydrocarbon chain (δ 1.213–1.328), and olefinic (δ 5.348, m) protons confirmed the existence of saturated and unsaturated fatty acids or their ester or amides in the chloroform fraction. The 1H NMR signals of GLJ-C also showed signals at δ 4.146 (dd, J = 6, 12 Hz) and δ 4.293 (dd, J = 4.2, 12 Hz), which represented the methylene group of the glycerol backbone from triglycerides [6,41].
Fig. 8.
1H NMR spectra of (A) GLJ-C and (B) GLJ-W/M. Numbers 1–15 represent the characteristic group peaks of compounds 1–15 in Table 1.
3.3.2. Characterization of the aqueous methanol fraction of GLJ
The 1H NMR spectrum of GLJ-W/M can be divided into three distinct regions (Fig. 8B). Organic acids and amino acids including threonine (δ 1.340, d, J = 7.2 Hz), acetic acid (δ 1.929 s), succinic acid (δ 2.454, s), succinate (δ 2.432, s), creatine (δ 3.059 s; δ 3.960, s), and glycine (δ 3.689, s) were identified in the corresponding characterized regions. Other signals (δ 3.216, s and δ 3.278, s; δ3.920, s) were identified as belonging to choline and betaine, respectively. Lactate (δ 4.134, q, J = 7.2 Hz), raffinose (δ 4.964, d, J = 4.2 Hz; δ 5.443, d, J = 3.6 Hz), and sucrose (δ 5.422, d, J = 4.2 Hz) were identified in the carbohydrate region, and formic acid (δ 8.462, s) was identified in the phenolic region. The chemical shifts and coupling constants of all the identified compounds are summarized in Table 1.
Table 1.
Chemical shift assignments in different fractions of GuiLingJi (GLJ).
| No. | Compounds | Signals | Fractions |
|---|---|---|---|
| 1 | Fatty acids or their ester or amides | 0.880 (t, J = 7.2 Hz), 2.346 (t, J = 7.2 Hz), 2.015 (m), 2.771 (t, J = 6.6 Hz), 1.213–1.328, 5.348 (m) | The chloroform fraction of GLJ (GLJ-C) |
| 2 | Triglycerides | 4.146 (dd, J = 6, 12 Hz), 4.293 (dd, J = 4.2, 12 Hz) | GLJ-C |
| 3 | β-rhamnosyl | 1.146 (d, J = 6 Hz, CH3) | The aqueous methanol fraction (GLJ-W/M) |
| 4 | Threonine | 1.340 (d, J = 7.2 Hz) | GLJ-W/M |
| 5 | Acetic acid | 1.929 (s) | GLJ-W/M |
| 6 | Succinate | 2.432 (s) | GLJ-W/M |
| 7 | Succinic acid | 2.454 (s) | GLJ-W/M |
| 8 | Creatine | 3.059 (s), 3.960 (s) | GLJ-W/M |
| 9 | Choline | 3.216 (s) | GLJ-W/M |
| 10 | Betaine | 3.278 (s), 3.920 (s) | GLJ-W/M |
| 11 | Glycine | 3.689 (s) | GLJ-W/M |
| 12 | Lactate | 4.134 (q, J = 7.2 Hz) | GLJ-W/M |
| 13 | Raffinose | 4.964 (d, J = 4.2 Hz), 5.443 (d, J = 3.6 Hz) | GLJ-W/M |
| 14 | Sucrose | 5.422 (d, J = 4.2 Hz) | GLJ-W/M |
| 15 | Formic acid | 8.462 (s) | GLJ-W/M |
3.4. Chemical composition of GLJ
An overview of UHPLC-QE HRMS and 1H NMR analyses combined with a polar division strategy for comprehensive characterization of the chemical composition of GLJ is shown in Fig. 9. To further clarify the distribution of the 150 compounds in different polar parts, a relative content heat map and Venn diagram were employed. Thus, only 82 compounds could be identified in GLJ-M; many components in the methanol extraction failed to be detected under the current test conditions due to serious signal interference. Moreover, 77 compounds were identified in GLJ-P, 114 compounds in GLJ-E, and 46 compounds in GLJ-B. More compounds were identified by using the method of polar division, and a total of 150 compounds were rapidly identified in all polar parts of GLJ by UHPLC-QE HRMS. In addition, 13 compounds including organic acids, amino acids, organic bases, and sugars were rapidly identified by 1H NMR spectroscopy. The 1H NMR spectra also definitively confirmed the existence of fatty acids, which were identified by UHPLC-QE HRMS.
Fig. 9.
The overview of the UHPLC-QE HRMS and 1H NMR for comprehensive characterization of chemical composition of GLJ.
3.5. Pharmacological activities of the main chemical components in GLJ
This study demonstrated that GLJ consists of multiple chemical components, which mainly include flavonoids and saponins, ginsenosides, coumarins, phenolic acids and so on. These chemical compositions have various pharmacological activities. For example, multiple pharmacological actions of ginsenosides have been observed to have anti-inflammatory activity and anti-central nervous system disorder effects, enhancing energy and sexuality [42]. In particular, ginsenosides Re and Rp1 possess anti-inflammatory properties by suppressing the NF-κB signaling pathway [43]. Ginsenoside Rb1 has an antifatigue effect by suppressing skeletal muscle oxidative stress and improving energy metabolism [44]. Ginsenosides Rb1 and Rg1 may significantly ameliorate learning and memory impairment by increasing the acetycholine level and inhibiting the decrease in 5-hydroxytryptamine in the brain [45]. In addition, ginsenosides Rb1 and Rg1 have been shown to improve testicular function in spermatogenesis by upregulating follicle stimulating hormone levels and luteinizing hormone levels [46]. Ginsenoside Rb1 also has a potent antifatigue effect caused by its suppression of skeletal muscle oxidative stress and improves energy metabolism [44]. A large number of flavonoids were identified in GLJ. Flavonoids can reverse age-related declines in neurocognitive performance by interacting with the cellular and molecular architecture of the brain responsible for memory and reducing neuronal loss due to neurodegenerative processes [47]. For example, epimedium flavonoids have been reported to improve learning-memory impairment and decrease the production of β-amyloid in the brains of amyloid-β precursor protein (APP) transgenic mice. These compounds might also improve cognitive impairment and white matter lesions induced by chronic cerebral hypoperfusion by inhibiting the Lingo-1/Fyn/ROCK signaling pathway and activating the BDNF/TrkB, NRG-1/ErbB4 and PI3K/Akt/CREB pathways in white matter [48]. Icariin, as the major ingredient of Epimrdii Folium, could improve learning and memory functions in APP/presenilin-1 transgenic mice, possibly by stimulating the NO/cGMP signaling pathway and coordinating the induction of nitric oxide synthase (NOS) isoforms [49]. Furthermore, it was reported to improve spatial learning and memory abilities as it causes a decreased expressions of tumor necrosis factor-α, interleukin-1, and cyclooxygenase-2 in the hippocampus [50]. In addition, phenolic acids such as caffeic acid have beneficial and/or protective effects in improving reproductive function, preventing sperm damage, ameliorating sperm function, and improving testicular function [51]. Coumarins have shown inhibitory effects against infection from various viruses such as human immunodeficiency virus, flu virus, enterovirus 71, and coxsackievirus A16 [52]. In conclusion, the synergistic combination of various compounds could achieve the multiple functions of GLJ. However, the current quality control standards of GLJ only include the content determination of two ginsenosides, which cannot fully reflect the quality of this formulation. The results of this study suggest that further attention should be paid to some flavonoids, which will help to find quality markers and improve quality standards.
4. Conclusion
In this study, a combined approach of UHPLC-QE HRMS and 1H NMR techniques coupled with a polarity partitioning strategy was used to comprehensively characterize multiple chemical components of GLJ. The compound database, CD software, diagnostic ion filtering, and mass defect filtering strategy were employed to analyze the mass spectrum. Finally, a total of 150 compounds were rapidly identified in GLJ by UHPLC-QE HRMS, of which 9 compounds were unambiguously identified by comparing the retention time and MS/MS data with those of reference compounds. In addition, 13 compounds were rapidly identified by 1H NMR spectroscopy. These two techniques could potentially complement each other. This analysis strategy can be applied to other TCM formulas for comprehensive characterization and identification of chemical components.
CRediT author statement
Jingchao Shi: Data curation, Writing - Original draft preparation; Xiaoxia Gao: Investigation, Writing - Reviewing and Editing; Airong Zhang: Data curation, Funding acquisition; Xuemei Qin: Supervision; Guanhua Du: Project administration.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We acknowledge financial support from the Shanxi Provincial Science and Technology Key Research and Development Plan (Grant Nos.: 201603D3113006 and 201903D311012). We also thank the Scientific Instrument Center of Shanxi University for the UHPLC-QE HRMS and 1H NMR analyses.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2021.09.013.
Contributor Information
Xuemei Qin, Email: qinxm@sxu.edu.cn.
Guanhua Du, Email: dugh@imm.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Pang H.Q., An H.M., Yang H., et al. Comprehensive chemical profiling of Yindan Xinnaotong soft capsule and its neuroprotective activity evaluation in vitro. J. Chromatogr. A. 2019;1601:288–299. doi: 10.1016/j.chroma.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 2.Pang B., Zhu Y., Lu L., et al. The applications and features of liquid chromatography-mass spectrometry in the analysis of traditional Chinese medicine. Evid. Based Compl. Alt. Med. 2016. 2016 doi: 10.1155/2016/3837270. 3837270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Z., Xu Y., Wang Q., et al. Metabolism and mutual biotransformations of anthraquinones and anthrones in rhubarb by human intestinal flora using UPLC-Q-TOF/MS. J. Chromatogr. B. 2019;1104:59–66. doi: 10.1016/j.jchromb.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Gao X.X., Wang N., Jia J.P., et al. Chemical profliling of Dingkun Dan by ultra high performance liquid chromatography Q exactive orbitrap high resolution mass spectrometry. J. Pharm. Biomed. Anal. 2020;177:112732. doi: 10.1016/j.jpba.2019.06.029. [DOI] [PubMed] [Google Scholar]
- 5.Xue Z.Z., Lai C.J.S., Kang L.P., et al. Profiling and isomer recognition of phenylethanoid glycosides from Magnolia officinalis based on diagnostic/holistic fragment ions analysis coupled with chemometrics. J. Chromatogr. A. 2020;1611:460583. doi: 10.1016/j.chroma.2019.460583. [DOI] [PubMed] [Google Scholar]
- 6.Yu S., Li J., Guo L., et al. Integrated liquid chromatography-mass spectrometry and nuclear magnetic resonance spectra for the comprehensive characterization of various components in the Shuxuening injection. J. Chromatogr. A. 2019;1599:125–135. doi: 10.1016/j.chroma.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Díaz-De-Cerio E., Aguilera-Saez L.M., Gómez-Caravaca A.M., et al. Characterization of bioactive compounds of Annona cherimola L. leaves using a combined approach based on HPLC-ESI-TOF-MS and NMR. Anal. Bioanal. Chem. 2018;410:3607–3619. doi: 10.1007/s00216-018-1051-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhao S.J., Tian J.S., Tai G., et al. 1H NMR-based metabolomics revealed the protective effects of Guilingji on the testicular dysfunction of aging rats. J. Ethnopharmacol. 2019;238:111839. doi: 10.1016/j.jep.2019.111839. [DOI] [PubMed] [Google Scholar]
- 9.Du K., Gao X.X., Feng Y., et al. Effects of Guilingji on Kidney-Yang deficiency syndrome in rats based on serum metabolomics. Acta Pharm. Sin. 2019;54:1476–1483. [Google Scholar]
- 10.Xie J., Wang J., Chen M., et al. Influence of Guilingji capsules on spermatogenesis and sexual hormones in oligospermatism rats. J. Guangzhou Univ. Tradit. Chin. Med. 2011;28:621–623. [Google Scholar]
- 11.Sun Z., Wu Y., Zou H., et al. Study on Guiingji capsule for mitochondria function of sperm in idiopathic asthenospermia. Chin. J. Androl. 2015;29:37–40. [Google Scholar]
- 12.Zhao S.J., Zhao X.Z., Liu H.L., et al. Effects of Guilingji on improving learning and memory dysfunction caused by aging. Chin. Tradit. Herb. Drugs. 2018;49:5352–5357. [Google Scholar]
- 13.Zhao S.J., Liu X.J., Tian J.S., et al. Effects of Guilingji on aging rats and its underlying mechanisms. Rejuvenation Res. 2020;23:138–149. doi: 10.1089/rej.2018.2118. [DOI] [PubMed] [Google Scholar]
- 14.Chinese Pharmacopeia Commission . Chinese Medical Science and Technology Press; Beijing: 2020. Pharmacopeia of the People's Republic of China; pp. 1055–1056. [Google Scholar]
- 15.Chu C., Xu S., Li X., et al. Profiling the ginsenosides of three ginseng products by LC-Q-TOF/MS. J. Food Sci. 2013;78:C653–C659. doi: 10.1111/1750-3841.12102. [DOI] [PubMed] [Google Scholar]
- 16.Wei H.L., Li Y.J., Chen J., et al. Triterpenoid saponins in roots of Achyranthese bidentata. Chin. J. Nat. Med. 2012;10:98–101. [Google Scholar]
- 17.Wu W., Sun L., Zhang Z., et al. Profiling and multivariate statistical analysis of Panax ginseng based on ultra-high-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2015;107:141–150. doi: 10.1016/j.jpba.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L.Q., Wang J., Li T., et al. Determination of the chemical components and phospholipids of velvet antler using UPLC/QTOF-MS coupled with UNIFI software. Exp. Ther. Med. 2019;17:3789–3799. doi: 10.3892/etm.2019.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song W., Qiao X., Chen K., et al. Biosynthesis-based quantitative analysis of 151 secondary metabolites of Licorice to differentiate medicinal Glycyrrhiza species and their hybrids. Anal. Chem. 2017;89:3146–3153. doi: 10.1021/acs.analchem.6b04919. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q., Ye M. Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice) J. Chromatogr. A. 2009;1216:1954–1969. doi: 10.1016/j.chroma.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 21.Luo J.M., Xiao X., Hong L., et al. Analysis on chemical constitutes in Psoraleae Fructus by combination of HPLC/TOF-MS and HPLC/IT-MSn. Chin. Tradit. Herb. Drugs. 2014;45:924–928. [Google Scholar]
- 22.Gan J.S., Ma Y., Wang Z.Y., et al. Analysis on chemical constituents in Epimedii Herba by UPLC/Q-TOF-MS. Drugs Clin. 2014;29:349–352. [Google Scholar]
- 23.Ma H.J., Gao J., Zhang Y.L., et al. Study on identifcation of compounds and their fragmentation pathways in licorice by HPLC-MSn. Chin. J. Tradit. Chin. Med. 2018;33:1120–1123. [Google Scholar]
- 24.Gao L. Comparative analysis of Flos Caryophyllata and Fructus Caryophylli by high-speed counter-current chromatography [master’s thesis] Dalian: LiaoNing Normal University. 2021 [Google Scholar]
- 25.Liu L.P., Xu X.F., Sun D.D., et al. Analysis on chemical components from water extract of Epimedii Folium by HPLC-ESI-Q-TOF-MS. Chin. J. Exp. Tradit. Med. Form. 2014;20:112–116. [Google Scholar]
- 26.Wang Y., Yuan L., Li Y.B., et al. Analysis on chemical constituents of Epimedii Folium by UPLC-Q-TOF-MS. Chin. Tradit. Herb. Drugs. 2017;48:2625–2631. [Google Scholar]
- 27.Montoro P., Maldini M., Russo M., et al. Metabolic profiling of roots of liquorice (Glycyrrhiza glabra) from different geographical areas by ESI/MS/MS and determination of major metabolites by LC-ESI/MS and LC-ESI/MS/MS. J. Pharm. Biomed. Anal. 2011;54:535–544. doi: 10.1016/j.jpba.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Farag M.A., Porzel A., Wessjohann L.A. Comparative metabolite profiling and fingerprinting of medicinal licorice roots using a multiplex approach of GC-MS, LC-MS and 1D NMR techniques. Phytochemistry. 2012;76:60–72. doi: 10.1016/j.phytochem.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Shakeri A., Masullo M., D'Urso G., et al. In depth chemical investigation of Glycyrrhiza triphylla Fisch roots guided by a preliminary HPLC-ESIMSn profiling. Food Chem. 2018;248:128–136. doi: 10.1016/j.foodchem.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 30.Li H., Wan L.R., Wang H., et al. Identification and mass spectrometric characterization of isomeric isoflavone aglycones by ESI-IT-TOF mass spectrometry. Chem. J. Chin. Univ. 2007;28:2284–2289. [Google Scholar]
- 31.Zhao Y.M., Liu S.X., Zhang C.X., et al. Analysis on chemical constituents from Glycyrrhizae Radix et rhizoma by HPLC-Q-TOF-MS. Chin. Tradit. Herb. Drugs. 2016;47:2061–2068. [Google Scholar]
- 32.Shan L., Yang N., Zhao Y., et al. A rapid classification and identification method applied to the analysis of glycosides in Bupleuri radix and liquorice by ultra high performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. J. Sep. Sci. 2018;41:3791–3805. doi: 10.1002/jssc.201800619. [DOI] [PubMed] [Google Scholar]
- 33.Yuan J., Gong Y.M., Ju P., et al. HPLC-MS2 analysis of chemical constituents in Epimedium koreanum. Chin. Tradit. Herb. Drugs. 2004;4:15–18. [Google Scholar]
- 34.Tong Y.X., Xu D.R., Kong L.Y. HPLC-MS3 analysis of chemical constituents in Epimedium koreanum, Chin. J. Nat. Med. 2006;4:58–61. [Google Scholar]
- 35.Zhu F.X., Zhao Y.G., Jia X.B., et al. Study on fingerprint of crude and processed Epimedium by UPLC-PDA-MS. Acta Chimica Sinica. 2012;5:635–642. [Google Scholar]
- 36.Yuan H., Cao S.P., Chen S.Y., et al. Analysis of 11 chemical constituents in Epimedium myrianthum Stearn by RRLC-DAD-ESI-MS2. Chin. J. Pharm. Anal. 2014;34:1156–1160. [Google Scholar]
- 37.Cheng Y., Wang N.L., Wang X.L., et al. Chemical constituents from Epimedium koreanum nakai. J. Shenyang Pharm. Univ. 2006;10:644–647. [Google Scholar]
- 38.Xie J., Zhang Y., Wang W., et al. Identification and simultaneous determination of glycyrrhizin, formononetin, glycyrrhetinic acid, liquiritin, isoliquiritigenin, and licochalcone A in licorice by LC-MS/MS. Acta Chromatogr. 2014;26:507–516. [Google Scholar]
- 39.Wang X.J., Zhu L.Z. Studies on the saponin constituents of Niu Qi (Achyrathes bidentata) J. Fourth Mil. Med. Univ. 1996;17:427–430. [Google Scholar]
- 40.Wang S., Zhu Y., Shao Q., et al. Identification of chemical constituents in two traditional Chinese medicine formulae by liquid chromatography-mass spectrometry and off-line nuclear magnetic resonance. J. Pharm. Biomed. Anal. 2016;117:255–265. doi: 10.1016/j.jpba.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Li A.P., Li Z.Y., Qu T.L., et al. Nuclear magnetic resonance based metabolomic differentiation of different Astragali Radix. Chin. J. Nat. Med. 2017;15:363–374. doi: 10.1016/S1875-5364(17)30057-2. [DOI] [PubMed] [Google Scholar]
- 42.Ratan Z.A., Haidere M.F., Hong Y.H., et al. Pharmacological potential of ginseng and its major component ginsenosides. J. Ginseng Res. 2021;45:199–210. doi: 10.1016/j.jgr.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D.H. Gut microbiota-mediated pharmacokinetics of ginseng saponins. J. Ginseng Res. 2018;42:255–263. doi: 10.1016/j.jgr.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan S.J., Zhou F., Li N., et al. Anti-fatigue effect of ginsenoside Rb1 on postoperative fatigue syndrome induced by major small intestinal resection in rat. Biol. Pharm. Bull. 2013;36:1634–1639. doi: 10.1248/bpb.b13-00522. [DOI] [PubMed] [Google Scholar]
- 45.Ahmed T., Raza S.H., Maryam A., et al. Ginsenoside Rb1 as a neuroprotective agent: A review. Brain Res. Bull. 2016;125:30–43. doi: 10.1016/j.brainresbull.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 46.Zhou S.H., Deng Y.F., Weng Z.W., et al. Traditional Chinese medicine as a remedy for male infertility: A review. World J. Mens Health. 2019;37:175–185. doi: 10.5534/wjmh.180069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer J.P., Vauzour D., Rendeiro C. Flavonoids and cognition: The molecular mechanisms underlying their behavioural effects. Arch. Biochem. Biophys. 2009;492:1–9. doi: 10.1016/j.abb.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Niu H.M., Wang M.Y., Ma D.L., et al. Epimedium flavonoids improve cognitive impairment and white matter lesions induced by chronic cerebral hypoperfusion through inhibiting the Lingo-1/Fyn/ROCK pathway and activating the BDNF/NRG1/PI3K pathway in rats. Brain Res. 2020;1743 doi: 10.1016/j.brainres.2020.146902. [DOI] [PubMed] [Google Scholar]
- 49.Jin F., Gong Q.H., Xu Y.S., et al. Icariin, a phosphodiesterase-5 inhibitor, improves learning and memory in APP/PS1 transgenic mice by stimulation of NO/cGMP signalling. Int. J. Neuropsychopharmacol. 2014;17:871–881. doi: 10.1017/S1461145713001533. [DOI] [PubMed] [Google Scholar]
- 50.Guo J., Li F., Wu Q., et al. Protective effects of icariin on brain dysfunction induced by lipopolysaccharide in rats. Phytomedicine. 2010;17:950–955. doi: 10.1016/j.phymed.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., Xu S.Y., Liu M.N., et al. Comparative studies on chemical contents and effect in Kidney-Yang deficiency rats of salt-processed product and wine-processed product of Cuscutae Semen. Evid. Based Complement Alternat. Med. 2019. 2019 doi: 10.1155/2019/2049497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mishra S., Pandey A., Manvati S. Coumarin: An emerging antiviral agent. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e03217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.