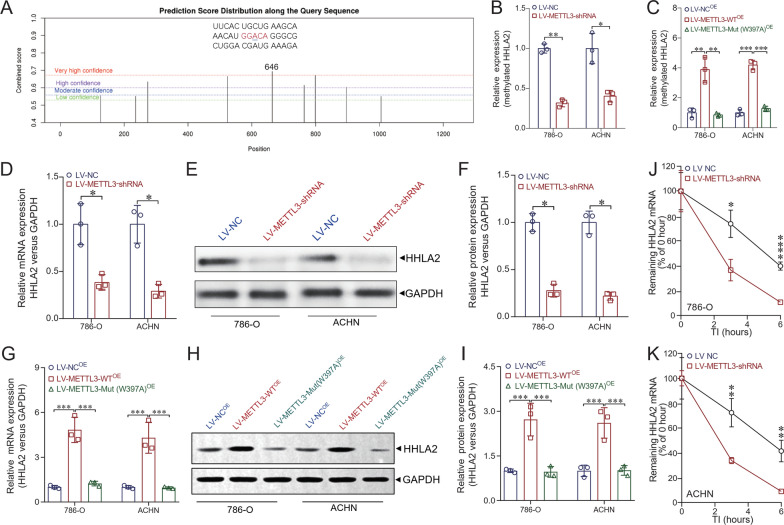
Fig. 3.
METTL3 regulates the expression of HHLA2 via m6A modification of HHLA2 mRNA. A The potential m6A site of HHLA2 mRNA was analyzed via a sequence-based m6A modification site predictor (SRAMP, http://www.cuilab.cn/sramp), and we found a very high confidence m6A site in HHLA2 mRNA (GGACA). B m6A-labeled mRNA level of HHLA2 was decreased in LV-METTL3-shRNA cells compared with LV-NC cells (P < 0.01 in 786-O and P < 0.05 in ACHN respectively). C m6A-labeled mRNA levels of HHLA2 were significantly elevated in METTL3-overexpressing ccRCC cells (P < 0.01 in 786-O and P < 0.001 in ACHN), but not in METTL3-Mut (W397A)-overexpressing ccRCC cells. D HHLA2 mRNA expression level was significantly decreased in LV-METTL3-shRNA compared with LV-NC in both 786-O and ACHN cells (P < 0.05). E and F Western blot analysis showed that HHLA2 protein expression level was significantly decreased in LV-METTL3-shRNA compared with LV-NC in both 786-O and ACHN cells (P < 0.05 respectively). G HHLA2 mRNA expression level was significantly increased in METTL3-overexpressing ccRCC cells (P < 0.001 in 786-O and ACHN respectively), but not in METTL3-Mut (W397A)-overexpressing ccRCC cells. H and I Western blot analysis showed that HHLA2 protein expression level was significantly increased in METTL3-overexpressing ccRCC cells (P < 0.001 in 786-O and ACHN respectively), but not in METTL3-Mut (W397A)-overexpressing ccRCC cells. J and K Changes of HHLA2 mRNA expression and decay rate after transcription inhibition were confirmed by qRT-PCR in the 786-O and ACHN cell lines when METTL3 was depleted. Un-paired t test was used as needed. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001