Figure 1.
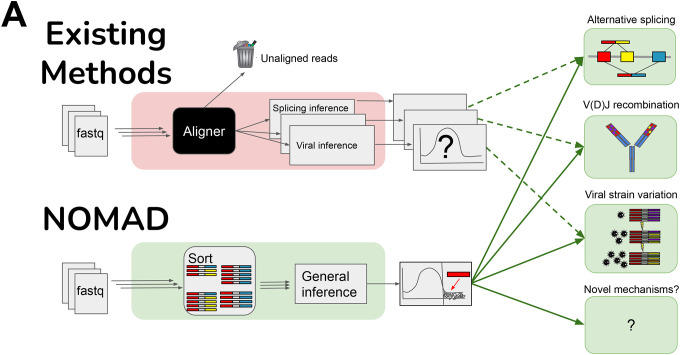
A. Overview of NOMAD (green) vs. existing methods (red). Typical workflows (red) remove reads during fastq preprocessing and alignment, and only then perform statistical significance testing. For every desired inferential task, a different inference pipeline must be used (red). NOMAD performs direct significance testing on raw fastq reads, bypassing alignment and enabling data-scientifically driven inference, using optional ontology mapping for interpretation. If optional mapping is desired, typically 1000 fold fewer reads than in initial fastqs files must be aligned.
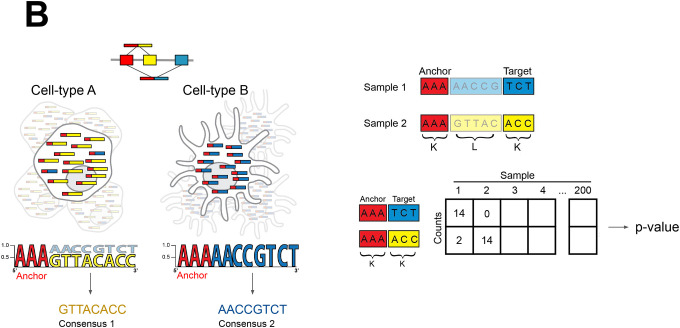
B. Overview of NOMAD statistics: raw fastq files are parsed into kmer anchors (red) and targets (blue and yellow) separated by a lookahead sequence of length L. For each anchor, statistical inference is performed on a contingency table of targets by samples. Reads with sample-dependent sequence diversification by alternative splicing are depicted. For each significant anchor, a per-sample consensus sequence is built which can be interpreted as the dominant isoform in the case of alternative splicing.
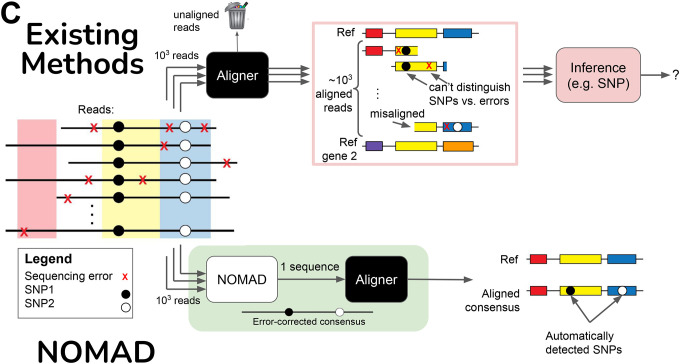
C. Consensus building denoises inputs to aligners before the alignment process. Sequencing errors (red X’s) are randomly distributed in reads, and by plurality vote across reads from the given sample, error-corrected as a consensus is built. Without this step, aligners will (a) fail to align, (b) yield misaligned reads, or (c) align reads correctly but with sequencing errors. Even if correct alignments are made, resulting mismatches with the reference must be further post-processed to make inference that discriminates sequencing errors from SNPs.
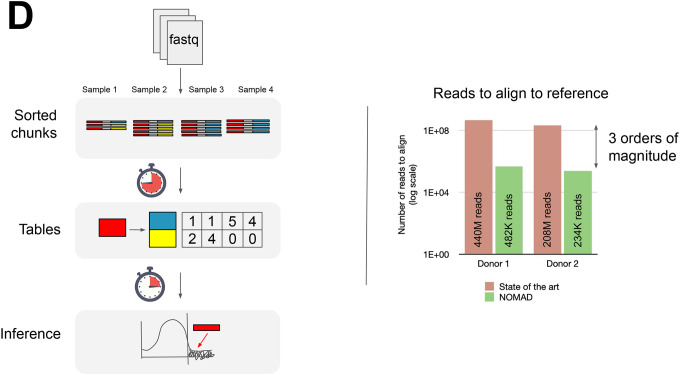
D. Left: NOMAD takes in fastq data, extracts (anchor, target) pairs of k-mers which are sorted and counted, and performs statistical inference. Right: After compressing and denoising via sample consensus sequences, NOMAD reduces the number of alignments required by a factor of 103.