Figure 2.
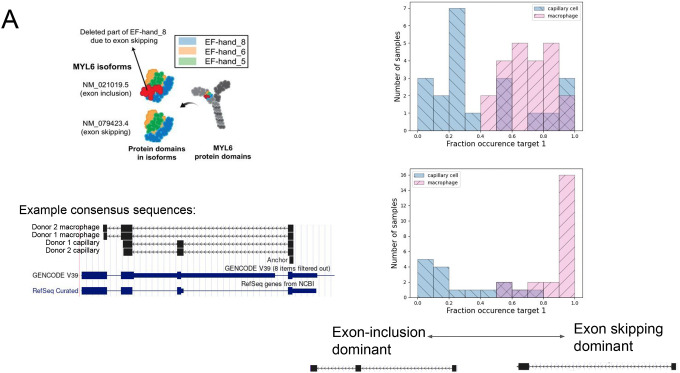
A. NOMAD detects anchors in MYL6, a positive control. Q value of 1.4E-8 for donor 1, 5.8E-41 for donor 2. Consensus split-read mapping shows capillary cells dominantly include and macrophage cells skip exon the exon in MYL6 including the EF_hand_8 domain (shown by the red color). Figure schematic taken from (36).
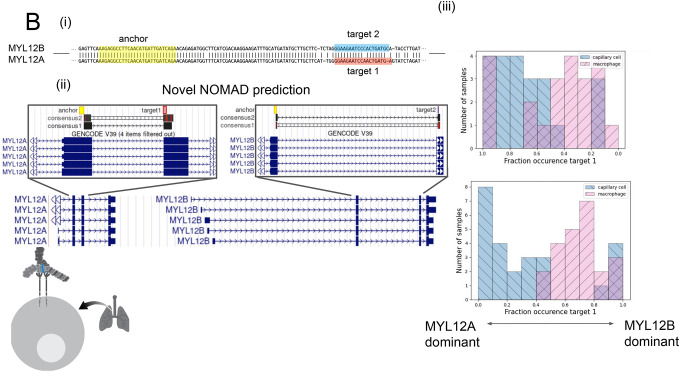
B. (i) Anchors mapping to MYL12 isoforms. Q value of 2.5E-8 for donor 1, 2.3E-42 for P3. An anchor highlighted in yellow includes a shared NOMAD-called anchor between donors 1 and 2; MYL12A and MYL12B isoforms share >95% nucleotide identity in coding regions. (ii) NOMAD’s approach automatically detects targets and creates consensus sequences that unambiguously distinguish the two isoforms. (iii) In both donors, NOMAD reveals differential regulation of MYL12A and MYL12B in capillary cells (MYL12A dominant) and macrophages (MYL12B dominant). The schematic illustrates MYL12 (in blue) within the myosin light chain complex, recently shown to interact with CD69 in the lung.
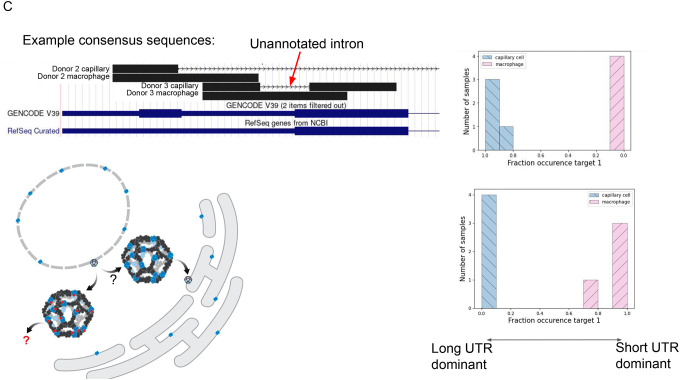
C. Schematic model shows SEC13 embedded in COP2-encased vesicles that bud from the endoplasmic reticulum; SEC13 is also found in the nuclear membrane. Short UTR isoforms, missing the C terminus of the protein, are dominant in capillary cells; long UTRs are dominant in macrophages. Each donor capillary cell has an identical consensus in donor 1, reflecting an unannotated splice isoform. Q value of 1.2E-9 for P2, 6.3E-26 for P3.
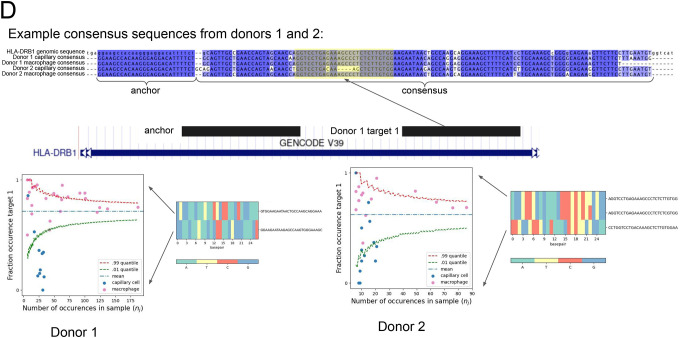
D. Unique alignment of the NOMAD-identified anchor, target1 pair to HLA-DRB1. Q value of 4.0E-10 for P2, 1.2E-4 for donor 2. Each donor, celltype pair has a distinct, cell-type specific consensus sequence, reflected by the multiway alignment to HLA-DRB1 3’ UTR. Nucleotide composition of the most abundant targets are depicted as heatmaps. Scatter plots show cell-type regulation of different HLA-DRB1 alleles not explained by a null binomial sampling model p<2E-16 for donor 1, 5.6E-8 for donor 2, finite sample confidence intervals depicted in red and green, statistical test described in Methods.
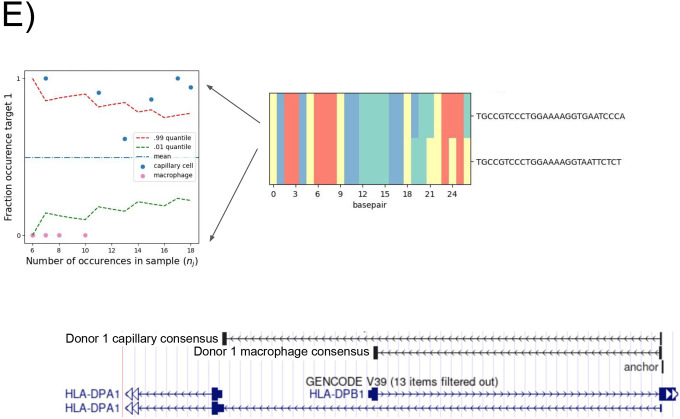
E. Donor 1 specific splice variant of HLA-DPB1. Anchor Q value: 7.9E-22. Detected targets in macrophages exclusively expressing the short isoform which shortens the ORF and changes the 3’ UTR; splice variants found de novo by NOMAD consensus’. Binomial hypothesis test as in D for cell-type target expression depicted in scatter plots (p<2.8E-14).