Figure 4: Unsupervised identification of V(D)J recombination in human and lemur immune cells.
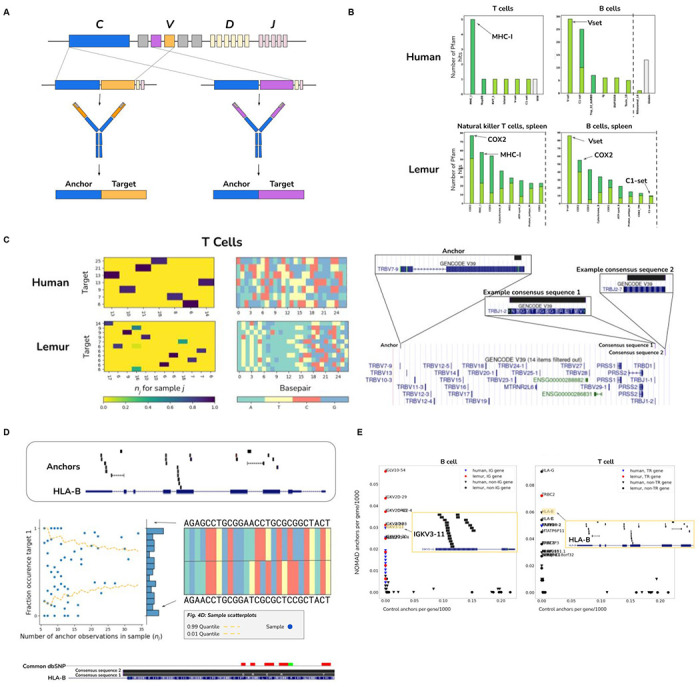
A. Stylized diagram depicting NOMAD detection of V(D)J recombination, with example variable regions in the heavy chain. An anchor sequence in the constant region (blue), generates target sequences (orange and purple) during V(D)J recombination, in which immunoglobulins may receive different gene segments during rearrangement. NOMAD is able to rediscover and detect these recombination events by prioritizing sample-specific TCR and BCR variants.
B. Unsupervised NOMAD protein profile analysis shows MHC and immunoglobulin variable regions are enriched in B and T cells. NOMAD recovers domains known to be diversified in adaptive immune cells, bypassing any genome reference or alignment; control hits computed from the most abundant anchors have no such enrichment. In B cells, hits in the V set, IG like domains resembling the antibody variable region, are at a relatively high E-value, as predicted by protein diversification generated during V(D)J, making matching to reference domains imperfect. The third most hit domain is Tnp_22_dsRBD, a double stranded RNA binding domain, suggesting potential activation of LINE elements in B cells. COX2, known to be involved in immune response, is highly ranked in both lemur T and B cells. Plots were truncated for clarity of presentation as indicated by dashed grey line (Fig. S2F–H).
C. NOMAD detects combinatorial expression of T cell receptors in immune cells de novo. In human T cells (right), we show a NOMAD anchor in the TRVB7-9 gene, and two example consensuses which map to disjoint J segments, TRBJ1-2 and TRBJ2-7. Histograms of this anchor depict combinatorial single-cell (columns) by target (row) expression of targets detected by NOMAD. Histogram for lemur T cells depicted similarly; lemur T cell anchor maps to the human gene TBC1D14.
D. NOMAD detects cell-type and allele-specific expression of HLA-B and HLA-B alleles de novo. NOMAD-annotated anchors are enriched in HLA-B (top Fig. 4.D.1). Sample scatterplot (middle)Fig. 4.D.2 shows that T cells have allelic-specific expression of HLA-B, not explicable by low sampling depth (binomial test as in Fig. 3d,e described in Methods, p< 4.6E-24). Fig. 4.D.3: HLA-B sequence variants are identified de novo by the consensus approach (bottom), including allele-specific expression of two HLA-B variants, one annotated in the genome reference, the other with 5 SNPs coinciding with annotated SNPs.
E. NOMAD analysis of lemur and human B (left) and T (right) cells recovers B, T cell receptors and HLA loci as most densely hit loci. Human genes are depicted as triangles; lemur as circles. Post facto alignments show variable regions in the kappa light chain in human B cells are most densely hit by NOMAD anchors and absent from controls; in T cells, the HLA loci and TRB including its constant and variable region are most densely hit, which are absent from controls. x-axis indicates the fraction of the 1000 control anchors (most abundant anchors) that map to the named transcript, y-axis indicates the fraction of NOMAD’s 1000 most significant anchors that map to the named transcript. Each inset depicts anchor density alignment in the IGKV region (left) and HLA-B in CD4+ T cells (top right) and TRBC-2 (bottom right), showing these regions are densely hit.
