Figure 4. B and T cell receptor diversity from human and lemur single cell data.
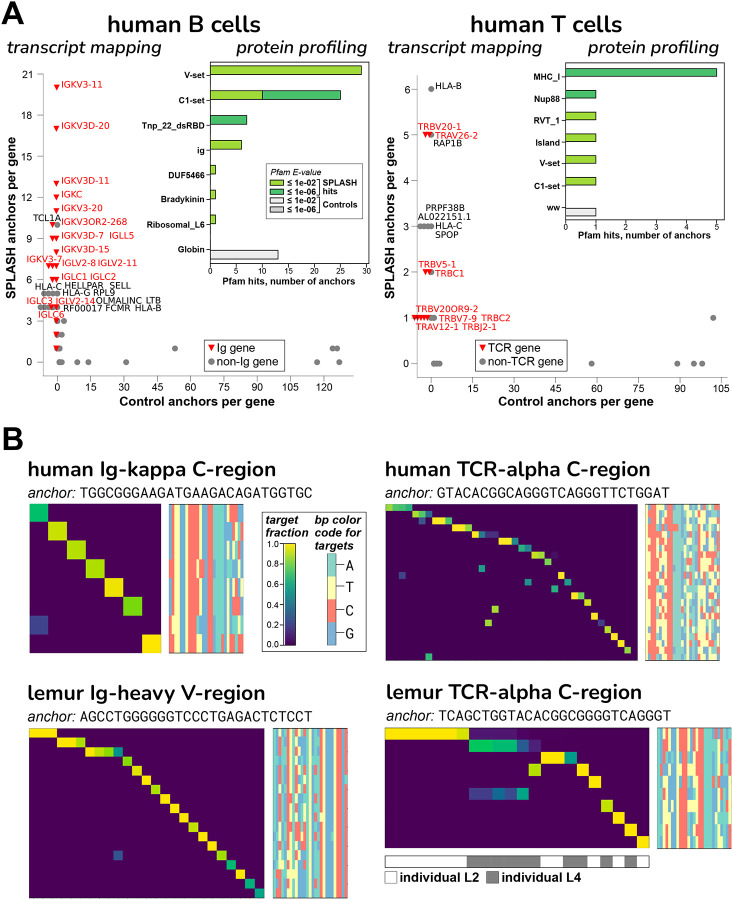
A. The “transcript mapping” plots show the number of anchors that align to a given gene name, for SPLASH on y-axis and Controls on x-axis, with immune receptor genes highlighted in red. For B cells, Ig genes (kappa = IGK and lambda = IGL) are by far the most predominant mapping among SPLASH anchors, but are not found at all in Control anchors (those with the highest counts). For T cells, TCR genes (alpha = TRA, beta = TRB) predominate, and are also not found in Controls. The inset histograms show that, in B cells and T cells, immunoglobulin-type “V-set” and “C1-set” are among the top protein domain annotations identified by Pfam on anchor consensuses, without using a reference genome. Mobile element activity is suggested by Pfam domains Tnp_22_dsRBD (“L1 transposable element dsRBD-like domain”) in B cells and RVT_1 (“Reverse transcriptase”) in T cells.
B. Targets associated with Ig/TCR anchors are clonotypically expressed, in both human and lemur: heatmaps show that most targets (rows) are expressed only in a single cell (columns). The anchors shown have among the highest target entropy scores for their gene type (Figure S5B illustrates an extreme example with 97 targets, for lemur Ig-lambda.) Target sequences are shown as bp color-maps (rows are targets, matching those in the heatmap; columns are bp positions, colored by base), giving a quick visualization of the sequence diversity. For lemur, NKT cells were studied, and show significant shared TCR usage – see top two rows; interestingly, the shared target sequence is different in the two individuals. There is similar sharing for lemur TCR-gamma and -beta chains (Figure S5C and D).