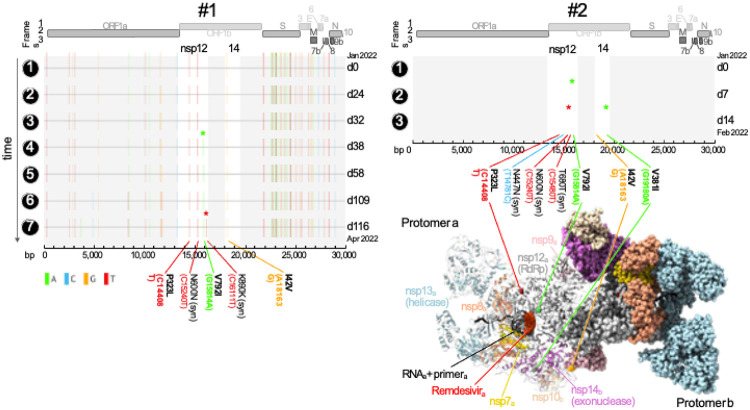
Figure 1.
Independent acquisition of the remdesivir resistance mutation nsp-V792I in two immunocompromised patients.
Full genome mutation profiles of SARS-CoV-2 viruses in longitudinal specimens of two immunocompromised patients treated with remdesivir. Basepair (bp) mutations compared to the Wuhan- Hu-1 reference are shown as ticks, color-coded according to the legend on the lower left. Bp and corresponding amino acid (aa) mutations in nsp12 (RNA-dependent RNA polymerase [RdRp]) and nsp14 (containing 3'-to-5' exoribonuclease proofreading activity) are labeled and shown in bold if non-synonymous. The longitudinal acquisition of mutations in nsp12 and 14 are highlighted by colored asterisks. Full genome maps are shown on top. The timeline is shown on the y-axis where time points are indicated on the left and days (d) elapsed since the first COVID-19 sampling per patient on the right of each plot.
A 3D protein structure of the multi-domain polymerase complex is shown in its active dimeric form. Each domain is colored differently and labeled in the protomer that is shown in ribbon representation, whereas the other domains are shown in sphere representation, respectively. Nsp14 (exonuclease activity) and its co-factor nsp10 convey RNA proofreading in trans and are thus highlighted/shown as ribbons in protomer b together with the other domains in protomer a. The non-synonymous mutations in nsp12 and 14 as well as remdesivir are highlighted and labeled. The structure of the polymerase complex dimer is based on pdb 7egq with remdesivir added by structural overlay of a remdesivir-bound nsp12 complex (pdb 7l1f)21,22.