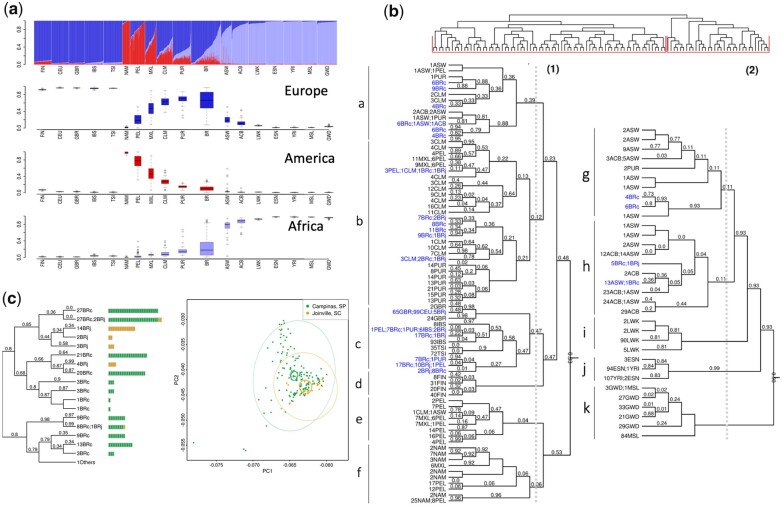
Fig. 1.
Admixture in Latino populations. a) ADMIXTURE analysis. Top panel: K = 3. Individuals are represented by vertical bars, the colors represent the estimated proportion of each cluster amounts to. Boxplots: genomic membership to each cluster: top: European ancestry; middle: Native American ancestry; and bottom: African ancestry. b) FineStructure tree of relationship across samples in the complete dataset. Posterior probability values below 1 are shown as branch labels. Dotted gray lines represent the cuts on the tree used to generate groups of distal clusters (a–k, see text). Edge labels show population membership and the number of individuals in each leaf. The dataset splits into 2 major clusters, according to non-African (1) and African (2) genomic predominance. Brazilians and Puerto Ricans are represented in both major clusters. Brazilians (in blue), however, are also more dispersed in distal clusters when compared to any other Latino Population, being the most dispersed admixed population, followed by ASW. BRc: samples from Campinas; BRj: samples from Joinville. c) Substructure in the Brazilian population. Left panel: FineStructure tree of relationship for Brazilian samples. Each colored bar corresponds to an individual, colors represent sample collection sites: green—Campinas (São Paulo State) and orange—Joinville (Santa Catarina State). Brazilian samples were subdivided into 18 leaves. Other populations were averaged and shown as super-individual (“1Others”). Right panel: PCA for PC1 and. PC2 on Brazilians. Inner ellipses are the 95% confidence ellipses for the barycenters of the groups. Outer ellipses are the 95% confidence ellipses for the groups.