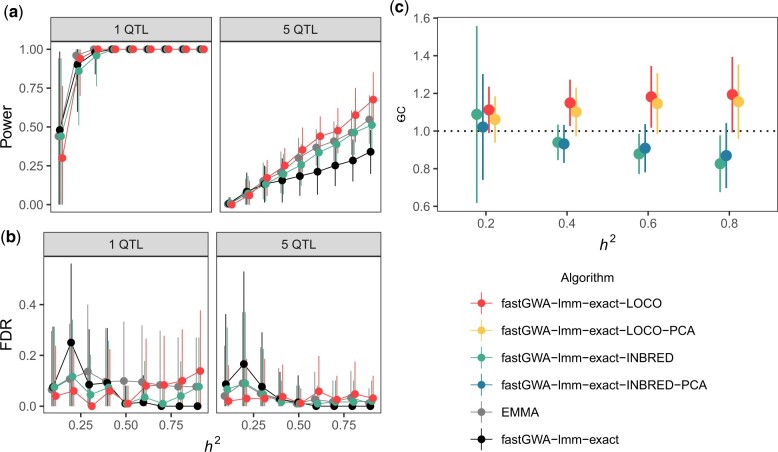
Fig. 1.
GCTA-based kinship matrix construction and GWAS outperforms EMMA. Estimates of power (a), and FDR (b), and genomic inflation (λGC) (c) of simulated mappings as a function of the narrow-sense heritability (x-axis) and the number of underlying QTL (strip titles). Simulations performed using EMMA, which undergirded cegwas2-nf (Zdraljevic et al. 2019), were compared with different formulations of kinship matrices and linear mixed model-based GWA implemented by GCTA (Yang et al. 2011; Jiang et al. 2019). In (c) metrics obtained from mappings that used indicated algorithms but also fitting the first eigenvector of the kinship matrix obtained from principal components analysis are also denoted.