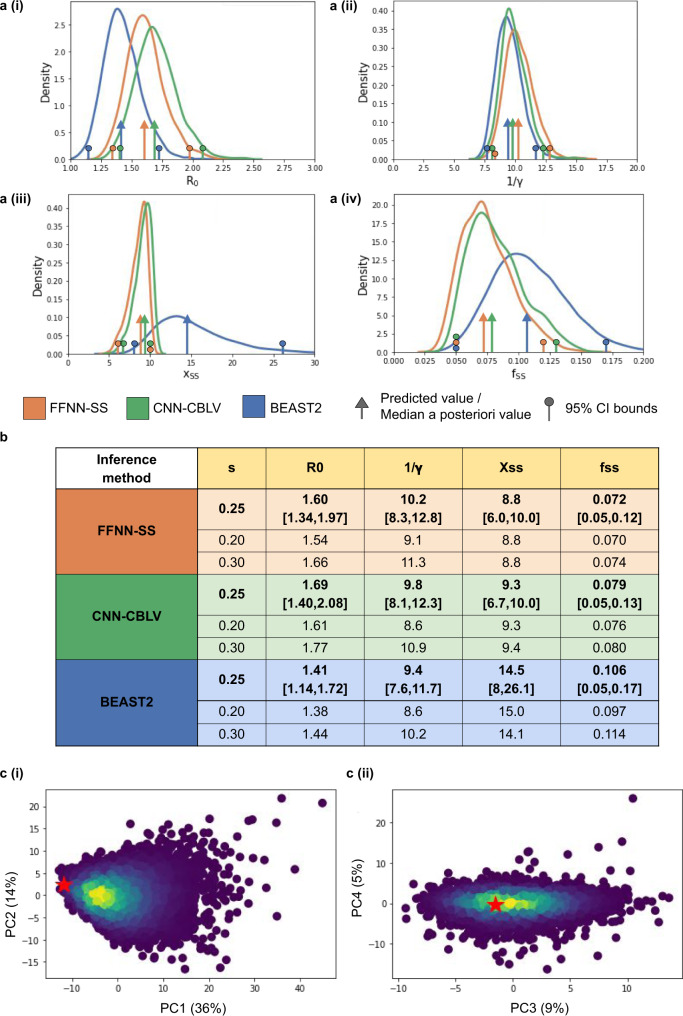
Fig. 5. Parameter inference on HIV data sampled from MSM in Zurich.
Using BDSS model with BEAST2 (in blue), FFNN-SS (in orange), and CNN-CBLV (in green) we infer: a (i) basic reproduction number, a (ii) infectious period (in years), a (iii) superspreading transmission ratio, and a (iv) superspreading fraction. For FFNN-SS and CNN-CBLV, we show the posterior distributions and the 95% CIs obtained with a fast approximation of the parametric bootstrap (‘Methods’, Supplementary Information). For BEAST2, the posterior distributions and 95% CI were obtained considering all reported steps (9000 in total) excluding the 10% burn-in. Arrows show the position of the original point estimates obtained with FFNN-SS and CNN-CBLV and the median a posteriori estimate obtained with BEAST2. Circles show lower and upper boundaries of 95% CI. b These values are reported in a table, together with point estimates obtained while considering lower and higher sampling probabilities (0.20 and 0.30). c 95% CI boundaries obtained with FFNN-SS are used to perform an a posteriori model adequacy check. We simulated 10,000 trees with BDSS while resampling each parameter from a uniform distribution, whose upper and lower bounds were defined by the 95% CI. We then encoded these trees into SS, performed PCA and projected SS obtained from the HIV MSM phylogeny (red stars) on these PCA plots. We show here the projection into c (i) first two components of PCA, c (ii) the 3rd and 4th components, together with the associated percentage of variance displayed in parentheses. Warm colours correspond to high density of simulations.