Abstract
Aedes albopictus is one of the most invasive insect species in the world and an effective vector for many important arboviruses. We reported previously that Ae. albopictus Nix (AalNix) is the male-determining factor of this species. However, whether AalNix alone is sufficient to initiate male development is unknown. Transgenic lines that express each of the three AalNix isoforms from the native promoter were obtained using piggyBac transformation. We verified the stable expression of AalNix isoforms in the transgenic lines and confirm that one isoform, AalNix3&4, is sufficient to convert females into fertile males (pseudo-males) that are indistinguishable from wild-type males. We also established a stable sex-converted female mosquito strain, AalNix3&4-♂4-pseudo-male. The pseudo-male mosquitoes can fly and mate normally with wild-type female, although their mating competitiveness is lower than wild-type. This work further clarifies the role of AalNix in the sex determination pathway and will facilitate the development of Ae. albopictus control strategies that rely on male-only releases such as SIT and sex-ratio distortion.
Author summary
Nix serves as a conserved male-determining factor in the two most important mosquito arboviral vectors, Ae. aegypti and Ae. albopictus. AaeNix alone can convert Ae. aegypti females into fertile but flightless males. AalNix has four alternative splice isoforms whereas AaeNix has one. Little was known previously about which AalNix isoform(s) serve as the primary signal for sex determination. We cloned the promoter region of AalNix gene and constructed piggybac-based AalNix overexpression constructs with different isoform variants. Following transformation and recovery of transgenic lines, we observed that expression of the AalNix3&4 isoform could shift the alternative splicing of the sex determination genes, doublesex and fruitless, from female to male isoforms, and phenotypically masculinize females or completely convert females into males. Importantly, the sex-converted pseudo-males are fertile and capable of flight. Thus, AalNix is the primary signal for male sex determination in Aedes albopictus and provides a basis for sex segregation and further Cas9-mediated gene-drive population suppression.
Introduction
Aedes albopictus is one of the most invasive and aggressive insect species in the world [1]. It is a competent vector for several globally-important arboviruses, including dengue (DENV), Zika (ZIKV), yellow fever and chikungunya (CHIKV) viruses [2–5]. Female mosquitoes require blood meals (hematophagy) from terrestrial vertebrates for both reproduction and nutrition. The process of blood feeding allows pathogens carried by the mosquitoes to be transmitted to vertebrate hosts and this poses a serious threat to human health [6].
In some Dipteran insects, including mosquitoes and non-Drosophilid flies, the presence of products encoded by a dominant male determining factor (M-factor) is the primary initiator of sex determination. Females in these species lack this factor and are designated m/m and males are heterogametic, M/m. The Nix gene was first identified as the M-factor in the yellow Fever mosquito, Aedes aegypti, and the AaeNix ortholog is linked to the 1st chromosome and is both required and sufficient to initiate male development [7]. Disruption of the Ae. albopictus ortholog, AalNix, can result in the feminization of males [8]. Guy1 and Yob/gYG2 are the candidate M-factors in the malaria vector mosquitoes, Anopheles stephensi and An. gambiae, respectively [9–11]. The Mdmd gene is the M-factor of the house fly, Musca domestica, and is indispensable for normal male development [12]. MoY is necessary and sufficient for male development and the M-factor of the fruit fly, Ceratitis capitata [13].
A cascade of genes in the sex-determination pathway relay the information from the primary signal to achieve terminal morphological and physiological differentiation [14]. In a previous study, we determined that the AalNix gene product is the primary signal for the Ae. albopictus sex-determination pathway [8]. However, whether AalNix alone can be used to initiate male determination requires further investigation. We also had shown that the male-specific doublesex gene isoform (dsxM) is the default splicing pattern in Aedes mosquito, which means that one or more potential dsx splicing enhancers link NIX and DSX [15]. The sex-specific splicing of dsx orthologues in Drosophila species is controlled by transformer (tra) [16–18]. Our working model is that accumulation of AalNix, a pre-mRNA splicing factor, is sufficient to regulate the alternative splicing of the unknown splicing enhancer, and cause this splicing factor to lose its ability to regulate dsx splicing, enabling dsx to recover its male-default splicing. However, no ortholog of tra has been found yet in mosquito families, most likely because it evolves rapidly resulting in significant sequence divergence [19,20]. The link of the M factor with the downstream dsx is still largely unexplored in mosquitoes.
Our previous work with AalNix showed that the gene has four exons (1–4) [8]. These exons are spliced alternately to produce four isoforms designated AalNix1 (contains all four exons), AalNix2 (exons 1, 3 and 4), AalNix3 (exon 1, intron 1 and 14 base-pairs (bp) of the 5’-end of exon 2) and AalNix4 (exon 1, intron 1 and 59 bp of the 5’-end of exon 2). AalNix1 has two regions homologous to the Ae. aegypti ortholog, AaeNix, that encode two RNA-binding motifs (RRM). AalNix2 encodes one RRM resulting from an exon 2 skip. AalNix3 and AalNix4 share the same stop codon due to retention of a short intron. Using mosquito transgenesis and expression constructs for each of the known AalNix isoforms, we show that AalNix3&4 alone is required and sufficient to convert females into fertile males with phenotypes similar to wild-type males. We also mated pseudo-males derived from genotypically m/m females carrying only the AalNix3&4 transgene to wild-type female mosquitoes and obtained stable lines. These studies support the further development of AalNix transgenes, as the male-determining factor of Ae. albopictus, for applications to sterile insect technologies (SIT) for mosquito control and provide the basis for the next step in the implementation of Cas9/guide RNA-based gene-drive systems in Ae. albopictus. Theoretically, gene-drive systems should result in the offspring of drive-system males and wild-type female mosquitoes being 50% gene-drive males, and the other 50% intersex or pseudo-males due to the introduced AalNix gene. The intersex do not produce offspring and pseudo-males, as good as the ones of line AalNix3&4-♂4 do produce some offspring including new pseudo-males, while wild-type males inheriting the gene-drive transgene have the same mating ability as wild-type males, and can mate with wild-type females to transmit the gene drive system resulting in population suppression. Our work confirms and extends a recent contribution by Lutrat et al [21].
Results
Generation of transgenic mosquitoes for each AalNix isoform
We made individual transgenes that linked the endogenous AalNix promoter to three distinct abbreviated forms of the coding region to see which of the four exons play roles in establishing male sex determination (Figs 1; S5). The transgenes were constructed to contain exons 1, 2, 3 and part of 4 (AalNix1), exons 1, 3 and part of 4 (AalNix2) or exon 1, intron1 and 59 bp of the 5’-end of exon 2 (AalNix3&4), and were inserted into a piggyBac transformation vector carrying a dominant fluorescent protein marker gene (OpIE2-DsRed). Following microinjection of embryos, matings of surviving injected adults (G0) to wild-type mosquitoes and screening of G1 progeny for the marker gene, three males and females each were recovered for AalNix1 (Table 1). The line derived from one of the males, AalNix1-♂2, was characterized further by inverse gene amplification procedures (iPCR) and shown to be inserted into an intergenic region at AaloF1:JXUM01S001974: 263598–263603 (S1A Fig). Three transgenic males and two females were recovered in the AalNix2 G1 progeny, and AalNix2-♂3, inserted intergenically at Aalbo_primary.1 scaffold_701: 209295–209300, was established for further analyses (S1B Fig). The G1 progeny arising from the AalNix3&4 transgenesis protocols included a number of mosquitoes with an intersex phenotype arising most likely from apparent conversion of females to males and providing preliminary evidence for the activity of this isoform combination in the sex-determination pathway (Table 1). We selected 21 phenotypically-normal males, six phenotypically-normal females expressing DsRed and four intersex individuals, and two lines, AalNix3&4-♂4 (Aalbo_primary.1 scaffold_64: 2942049–2942054) and AalNix3&4-♂15 (Aalbo_primary.1 scaffold_1: 15815977–15815982), were established for further analyses (Figs 2; S1C). Both of these insertions were intergenic based on the genome annotation and did not disrupt any coding sequence, and the insertion site of AalNix3&4-♂4 is shown in (Figs 2C; S1). Nonetheless, we do not rule out the possibility that inverse PCR cannot detect all transgene insertions in a transgenic line.
Fig 1. Donor plasmids used to generate transgenic Ae. albopictus expressing AalNix isoforms.
All constructs shown in the figure are flanked by the piggyBac arms to facilitate transposon-mediated integration into the Ae. albopictus genome. The DsRed fluorescent marker gene under the control of the OpIE-2 promoter serves as a dominant marker gene for transformation. AalNix isoform variants were designed to be expressed by the AalNix promoter. The C-tag refers to the Strep tag II placed at the carboxyl terminus of the AalNIX protein.
Table 1. Embryo microinjection results and screening of G1 progeny for Nix transformants.
| Donor plasmid | # embryos injected | #G0 survivors (%) | # pools | # of G1 screened | Pools with DsRed G1 progeny (#) |
|---|---|---|---|---|---|
| AalNix1 | 1,150 | 446 (38.78%) | 3 | 19,404 | P3: (3♂; 3♀) |
| AalNix2 | 895 | 148 (16.54%) | 2 | 9,146 | P1: (3♂; 2♀) |
| AalNix3&4 | 1,073 | 150 (13.98%) | 2 | 10,675 | P2: (21♂; 6♀; 4 intersex) |
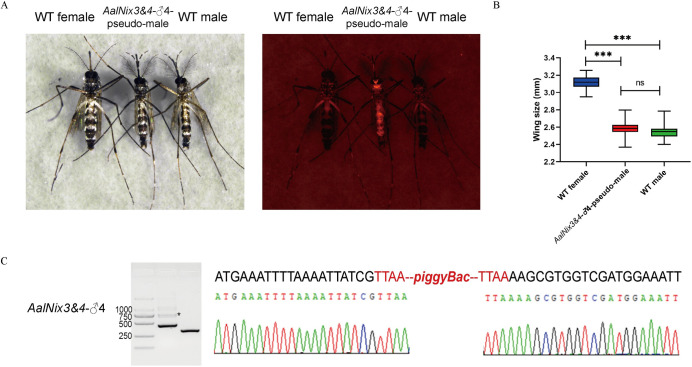
Fig 2. The AalNix3&4 transgene alone is sufficient to convert genetic females into fertile males.
(A) The left panel is the bright-field image of the three genotypes of WT female, AalNix3&4-♂4-pseudo-male and WT male. The right panel is the florescent image of the same individuals. (B) Average length of the left and right wings of individuals of the three groups (WT female, AalNix3&4-♂4-pseudo-male and WT male). Thirty individuals within each group were measured from the same cohort. The box plot, starting from bottom, shows minimum values, first quartile, median, third quartile, and maximum values using horizontal solid lines, with the mean indicated by a horizontal line. (C) Analysis of the insertion site of transgenic AalNix3&4-♂4 strain. Agarose gel electrophoresis of PCR products showing the results of amplifying the flanking regions. Sequence analysis showed that and the insertion site is in intergenic region. The signal marked by the asterisk (*) results from incomplete digestion.
AalNix3&4 is sufficient alone to convert females into males
We designed two pairs of oligonucleotide primers and used gene amplification analyses (PCR) to explore the inheritance profiles of the transgenic AalNix isoforms (S1 Table; S2A Fig). The first primer pair Nix_E1F/strep tag-R spans exon 1 and an inserted Strep II tag, and was designed to amplify the transgene insertion. The second primer pair, Nix_i2-F/Nix_i2-R, located in intron 2, are designed to distinguish endogenous AalNix from transgenic Nix (S2B Fig). Amplicons corresponding to the inserted AalNix1 and AalNix2 transgenes were detected in AalNix1-♂2and AalNix2-♂3, respectively, but no transgenic individuals exhibited sex conversion or intersex phenotypes (S2 and S3 Tables).
In contrast, all genetic female mosquitoes (m/m) of the AalNix3&4-♂4 line (confirmed by gene-specific [native vs transgenic] PCR) carrying the transgene (Nix presence indicated by DsRed) display only sex-converted male or intersex phenotypes (Fig 2; S4 Table). Expression of AalNix3&4 at a site distant from the M locus results in an intersex phenotype characterized by incomplete and/or deformed male-specific, external reproductive organ-like structures in the terminal abdominal segments (S3 Fig). Interestingly, dissections of the intersex individuals revealed the presence of male-specific reproductive organs, testis and accessory glands, with abnormal morphology (S3 Fig).
The pseudo-males (m/m; AalNix, DsRed+) display complete and normal-appearing male-specific external and internal reproductive organs including gonocoxites, gonostyli, testes and accessory glands. In addition, they display the plumose antennae characteristic of wild-type males (Figs 2; S3). We selected randomly a G2 AalNix3&4-♂4 pseudo-male, mated it with wild-type females and established a stable line, AalNix3&4-♂4-pseudo-male, that has been propagated for >17 generations and remains stable. Remarkably, there is a complete absence of an M-locus in this cross, which was thought previously essential for the sexual development of male mosquitoes [8]. The wing-length measurements of the pseudo-male offspring of this line are comparable in size to wild-type males, supporting the conclusion that the converted males are indistinguishable from their wild-type counterparts unless viewed with fluorescence microscopy, where they display the DsRed phenotype (Fig 2B; S5 Table). All of the pseudo-males recovered in subsequent generations (G3-G10) are DsRed+, while the females are not, and the sex-ratio is ~1:1 (p = 0.865) (S6 Table). These data confirm that the AalNix3&4 isoform is sufficient alone to convert genotypic females (m/m) to fully phenotypic males.
The AalNix3&4-♂15 line yielded four phenotypes during its establishment, normal females, intersexes, converted and the wild-type males (S7 Table). The differences in the characteristics of this line when compared to AalNix3&4-♂4 could result from insertion site effects from it being integrated at a different location in the genome. Also, there were multiple piggyBac transgene insertions in these lines, and some of these insertions (at least one) were not masculinizing and produced transgenic females. Furthermore, genotypically-female (m/m), transgenic (Nix, DsRed+) converted males were rare (~0.5%, 21 of 4102) and the strain went extinct at 10 generations.
Transgene-expressed AalNix alters doublesex (dsx) and fruitless (fru) splicing
Endogenous AalNix expression is first detected in embryos between 4h and 8h after oviposition and thereafter throughout subsequent life stages. Importantly, transcription of AalNix is limited to male mosquitoes and regulates the splicing of the zygotic AaldsxM isoform [8]. We investigated the impact of exogenously-supplied AalNix on dsx and fru to determine its impact in regulating the sex determination pathway in the transgenic mosquitoes.
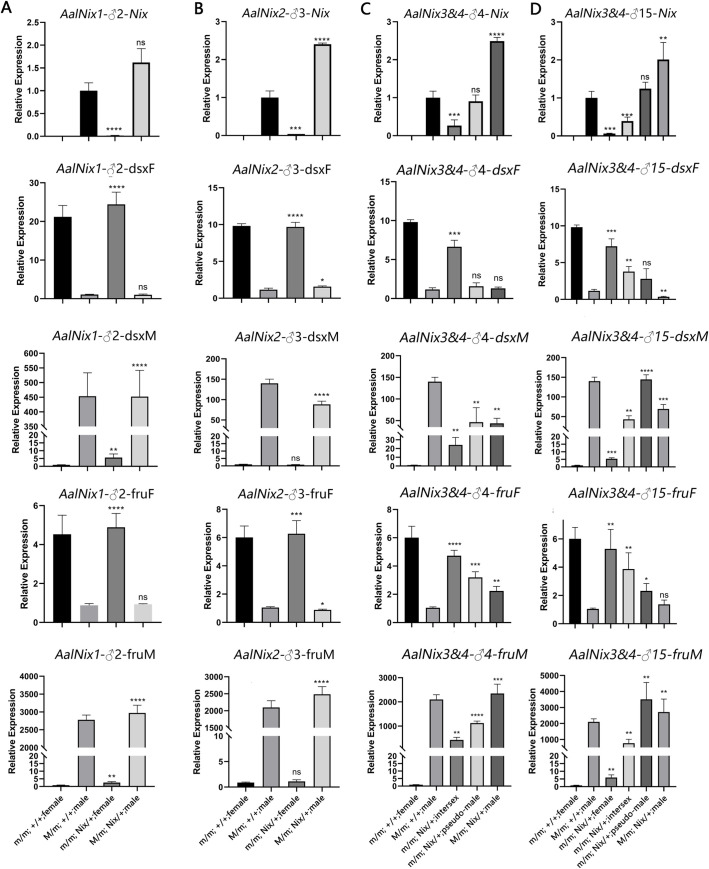
AalNix expression in AalNix1-♂2 and AalNix2-♂3 transgenic males was 1.62-fold and 2.40-fold higher than wild-type males, respectively, while as expected, AalNix transcripts are not found in transgenic females (Fig 3). As a downstream gene of sex determination pathway, the female and male isoforms of dsx and fru do not change in the AalNix transgenic individuals, compared to wild-type individuals (Fig 3).
Fig 3. Quantification (qRT-PCR analysis) of relative mRNA levels of AalNix and downstream genes in wild-type males and females, and each phenotypic of transgenic mosquito in each strain.
AalNix, Aalbdsx and Aalfru of AalNix1-♂2(A), AalNix2-♂3 (B), AalNix3&4-♂4 (C) and AalNix3&4-♂15 (D). The results were normalized to the Ae. albopictus Rps7 (AalRpS7) gene and are shown as the mean ± SD of three technical replicates.*P<0.05;**P<0.01; ***P<0.005; ****P<0.0001.
The expression levels of AalNix gene transcripts in AalNix3&4-♂4 (carrying the extra copy of the expressed first exon and 24 bp of intron 1) intersex individuals, pseudo-males and male were 0.27-, 0.9- and 2.48-fold greater, respectively, than wild-type males (Fig 3A–3D). The female-specific dsxF splice variant was the main isoform detected in wild-type females, and the male-specific splicing variant, dsxM was predominant in males. Interestingly, dsxF transcripts are abundant in intersex mosquitoes, which are phenotypically more similar to wild-type females. Furthermore, the transcript accumulation of dsxF in pseudo-males is 0.16-fold lower than wild-type females and is closer to that of wild-type males. The transcript accumulation levels of the dsxM isoform in intersexes is 17% that of wild-type male mosquitoes and 24-fold higher than of wild-type females. However, dsxM accumulates to similar levels in pseudo-males and genotypic males (M/m) carrying the AalNix3&4 transgene and these are lower than in wild-type males. The expression trends of fruF and fruM are similar to dsxF and dsxM (Fig 3A–3D).
Mating competitiveness of transgenic male mosquitoes
Competition assays in cages involved exposing ten wild-type females to mixed groups of pseudo-males, transgenic males expressing both native AalNix and exogenous AalNix3&4 and wild-type males. Following exposure to a 1:1 ratio of five pseudo-males to five wild-type males, the percentage of females that produced broods that contained transgenic progeny was 15% (9 of 51) (Table 2) and the percentage of transgenic progeny was 9.08% (251 of 2,512) (S8 Table). Although the visible morphological phenotypes of the transgenic males are not distinguishable from those of wild-type males (Fig 2), they contribute significantly fewer progeny than the wild-types to the next generation (p < 0.001; Table 3).
Table 2. Proportion of females of the AalNix3&4-♂4 line that produced broods with transgenic progeny.
| Brood with transgenic progeny | Brood with no transgenic progeny | Observed proportion(%) | |
|---|---|---|---|
| Exp11 (m/m; Nix/+) pseudo-male: (M/m; +/+) WT♂ = 1:1 | 9 | 51 | 15.0% |
| Exp22 (M/m; Nix/+) male: (M/m; +/+)WT♂ = 1:1 | 35 | 25 | 58.3% |
1. 5 pseudo-male mosquitoes mixed with 5 WT male, and then mated with 10 unmated WT female mosquitoes
2. 5 transgenic male mosquitoes mixed with 5 WT male, and then mated with 10 unmated WT female mosquitoes
Table 3. Proportion of transgenic progeny in AalNix3&4-♂4 line competition experiments.
| Replicate | DsRed+ | DsRed- | Percent |
|---|---|---|---|
| 1 | 25 | 436 | 5.42 |
| 2 | 34 | 398 | 7.87 |
| 3 | 31 | 403 | 7.14 |
| 4 | 77 | 543 | 12.42 |
| 5 | 59 | 424 | 12.22 |
| 6 | 25 | 308 | 7.51 |
| Total | 251 | 2512 | 9.08 |
Five AalNix3&4- ♂4 pseudo-male (m/m, ♂) compete with five wild-type males (M/m).
*(p < 0.001)
A competition experiment in cages (six replicates) involving exposing ten wild-type females to a 1:1 ratio of wild-type: heterozygous transgenic males (M/m; Nix/+) resulted in 58.3% (35 of 25) (Table 2) of the females producing broods with transgenic progeny of which 31.89% (603 of 1,288) (p < 0.001) were transgenic (Table 4; S9 Table). This data support the further development of these or similar transgenic strains for application to future population replacement or population suppression genetic control strategies.
Table 4. Proportion of transgenic progeny.
| Replicate | DsRed+ | DsRed- | Percent (%) |
|---|---|---|---|
| 1 | 86 | 247 | 25.83 |
| 2 | 94 | 271 | 25.75 |
| 3 | 144 | 180 | 44.44 |
| 4 | 93 | 225 | 29.25 |
| 5 | 95 | 200 | 32.20 |
| 6 | 91 | 165 | 35.55 |
| Total | 603 | 1288 | 31.89* |
Five AalNix3&4-♂4 male(M/m, ♂) compete with five wild type males.
*(p < 0.001)
Discussion
The results of these experiments demonstrate that the expression of exogenously-derived AalNix gene products can convert genotypically (m/m) Ae. albopictus females into males. Transgenic females carrying and expressing the first AalNix exon can develop into functional adult males capable of mating with wild-type females to produce normal offspring. These data are consistent with a recent report and confirm that the Nix gene alone is sufficient to initiate male development in Ae. albopictus, and lays the foundation for the next step to link the Nix gene with a CRISPR/Cas9 gene-drive system for control of vector-borne diseases.
Aedes albopictus and Ae. aegypti have a sex-determining locus, M, on their 1st chromosomes. This chromosome behaves generally in all other aspects as an autosome. There are two annotated protein-encoding genes in the Ae. aegypti M-locus, including Nix, and a myo-sex gene that has a role in the flight of adult males [22]. However, only one gene orthologous with AaeNix has been found in the M-locus of Ae. albopictus and it is designated AalNix [23]. Unlike AaeNix, which has only a single isoform, AalNix has four isoforms (AalNix1-4). The coding sequence of AalNix3 and AalNix4 are nearly identical and contain only the first RRM motif found in AaeNix [8]. Intron 1 and its 5’-end donor site location is conserved among AaeNix and AalNix (S4 Fig). It is worth noting that AalNix3 and AalNix4 differ from other isoforms in that their CDS includes 24 bases of the AalNix gene intron1. As an RRM motif-containing protein, NIX is critical for dsx and fru male-specific transcript splicing in the sex-determination pathway. We constructed three AalNix isoform expression plasmids and were able to establish the transgenic strains of each isoform. Expression of exogenously-provided AalNix1 and AalNix2 did not result in sex conversion in the females in the corresponding transgenic lines, but we did not recover a larger number of lines. In contrast, genetic females (m/m) of AalNix3&4 lines showed conversion to morphologically-normal wild-type (M/m) males. The different AalNix3&4 insertion sites in two distinct transgenic lines produced genetic females (m/m) with different degrees and proportions of male conversion, most likely resulting from an insertion-site effects. In addition, it is known that using piggyBac is likely to produce multiple-copy insertions [24], and inverse PCR mapping in lines most-likely detected only one of the multiple insertions, not necessarily the masculinizing one, results in the differences observed between AalNix3&4-♂4 and AalNix3&4-♂15. As seen with previous work with AalNix gene [8], the AalNix3&4 isoform was found in male pupae, male adults and C6/36 cells, supporting the conclusion that it plays a major role in the development of sex characteristics of Ae. albopictus males. While it cannot be ruled out that the insertion-sites affect AalNix1 and AalNix2 function because both contain exon 1, AalNix3&4 has the extra 25 bp of intron 1 and contains only one RRM site. It is possible that AalNix1 and AalNix2 may have roles assisting AalNix3&4 in the male sex determination pathway. As shown in the mating competition experiment, wild-type males carrying the transgene were as competitive as those without the transgene, but the mating competitiveness of pseudo-males was reduced.
We also isolated a completely gender-converted female from the AalNix3&4-♂4 strain, and following mating it with wild-type females to establish a stable strain, all subsequent male progeny of outcrosses to wild-type females inherited the AalNix3&4 transgene (DsRed+), while the females did not (DsRed-). Thus, we have inserted an artificial M-locus into the female (m/m) genome and produced fully-functional males through sex conversion.
The strain carrying the male sex-linked fluorescent markers are ideally suited for automated fluorescence sex-sorting needed for technologies such as SIT where it is important to obtain large numbers of male mosquitoes. Furthermore, since Nix alone completely converts females to males, combining it with a gene-drive system could produce a strain in which all resulting progeny are males following outcrossing [19,25]. We are working now to build such a gene-drive system in Ae. albopictus based on gender conversion and this could help achieve the goal of eliminating breeding female mosquitoes by producing pseudo-male mosquitoes.
Materials and methods
Mosquitoes
The Ae. albopictus Foshan strain was a kind gift from the Centers for Disease Control of Guangdong Province. This strain was isolated from Foshan, Guangdong, PRC and has been established in the laboratory since 1981. All mosquitoes are reared in an insect breeding room with constant temperature and humidity (27 ± 1°C, relative humidity 70%-80%) under a 14 L:10D photoperiod. The larvae were reared in plastic pots and fed ground turtle food (INCH-GOLD, Shenzhen, China). Adult mosquitoes were kept in 20 X 20 X 30 cm nylon mesh cages and allowed access to a cotton wick soaked in a 10% glucose solution and mated freely. Adult females were allowed to take blood meals from defibrinated sheep blood (Solarbio Life Sciences, Beijing, China) using a Hemotek membrane feeding system (Discovery Workshops, Accrington, UK).
Plasmid construction
All oligonucleotide primers are listed in S1 Table. To generate the pAalNix constructs, the piggyBac backbone plasmid AAEL010097-Cas9 (a kind gift from Omar Akbari [Addgene plasmid # 100707; http://n2t.net/addgene:100707]) [3] was digested and ligated to a linearized AalNix cassette (containing the CDS of AalNix1, AalNix2, AalNix3) at Not I/Asc I sites using the In-Fusion HD Cloning Kit (Takara, Dalian, China) [26]. To construct the piggyBac-AalNix1 plasmid, a 3,967-bp sequence containing the 2,478-bp AalNix native promoter (S5 Fig) (including the 133-bp 5′-end UTR), the 999-bp Nix1 ORF (the 24-bp Strep-tag II fusion to the C-terminus of AalNix1), and a 239 bp 3′-end UTR region followed by the bGH poly(A) signal, was amplified from pAalNix1 [15] using the primers Aal01 and Aal02 and cloned into the linearized backbone vector described above. To construct the piggyBac-AalNix2 plasmid, the backbone of the AAEL010097-Cas9 Vector and two DNA fragments were then joined in the following order: 1) AalNix native promoter and exon1 sequence (674bp) was amplified from pAalNix1 using the primer Aal03 and Aal04; 2) the sequence of the remaining 247-bp AalNix2 ORF region (Strep-Tag II fusion to the C-terminus of AalNix2) and 159 bp 3′-end UTR region followed by bGH poly(A) signal (this fragment was synthesis in vitro (Invitrogen, Guangzhou, China) using primers Aal05 and Aal06. These two fragments were purified and joined using the In-Fusion HD Cloning Kit (Takara), resulting in the pAalNix2 plasmid. The pAalNix3&4 plasmid was constructed using the same method for the pAalNix2 plasmid: 1) the AalNix native promoter and exon1 sequence were amplified from pAalNix1 using the primer Aal07 and Aal08; 2) the sequence of the remaining 25-bp AalNix4 ORF region (Strep-Tag II fusion to the C-terminus of AalNix4) and 138 bp 3′ UTR region followed by bGH poly-(A) signal sequence (Invitrogen Gene Synthesis service) using primers Aal09 and Aal10. These two fragments were purified and inserted into pAalNix3&4 plasmid using the In-Fusion HD Cloning Kit (Takara).
All amplification steps were performed using Q5 Hot Start High-Fidelity 2X Master Mix (NEB, Ipswich, MA) and the amplicons were analyzed by agarose gel electrophoresis and, then purified using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific, Carlsbad, CA, USA). All pAalNix plasmid sequences were verified using Shenggong Biotech Sanger sequencing services (Shenggong Biotechnology Co. Ltd., Guangzhou, China). Plasmids with the correct sequence were used as donors for construction of transgenic mosquitoes.
Generation of Ae. albopictus transgenic lines
The injected plasmid DNA mixture comprised a construct donor plasmid (500 ng/μL) and the phsp-Pbac helper plasmid [19,27,28] (300 ng/μL, a kind gift from Anthony A. James) mixed in injection buffer [29] and co-injected into 0-1h old embryos [30]. After microinjection, 16-20h old injected egg were heat shocked at 37°C for 1h, and then placed at 28°C, 80% humidity until hatching.
Characterization insertion site of the transgenic line
Inverse PCR was performed to characterize the insertion site as described previously [3,22]. Genomic DNA was extracted from adult females and males from each line, using a DNeasy Blood and Tissue kit (Qiagen, Hinden, Germany) according to the supplier’s protocol. One microgram of DNA was digested for 8h at 37°C with Sau3AI (5’-end reaction) or HinP1I (3’-end reaction) restriction enzymes (NEB), and then purified using the GeneJET PCR Purification Kit (Thermo Fisher Scientific) in accordance with the manufacturer’s protocol. Approximately 200 ng DNA fragments were circularized with T4 DNA ligase (NEB) at 16°C overnight in a 200 uL reaction volume. Finally, the ligation product was purified and subjected to two rounds of PCR reaction using Q5 Hot Start High-Fidelity 2X Master Mix (NEB) with primers AE049-AE056 [3]. Primers Aal049/ Aal050 and primers Aal053/ Aal054 were used for the first round of PCR for 5’ -end reaction and 3’-end reaction respectively. Primers Aal051/ Aal052 and primers Aal055/ Aal056 were used for nested PCR for 5’-end reaction and 3’-end reaction, respectively. The nested PCR products were directly loaded on agarose gels, and were extracted using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific), ligated using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific), and transformed into DH5α competent cells (Takara). Positive clones were purified (EZNA Plasmid Mini Kit I, Omega) and sent for sequencing (ShengGong Biotechnology Co. Ltd., Shanghai, China).
Identification of endogenous and transgenic AalNix
To ensure that all transgene cassettes were stably inherited to the next generation, male phenotype individuals with DsRed fluorescence were selected and a part of the leg cut off to extract DNA, using the Extract-N-Amp Tissue PCR Kit (Sigma-Aldrich Inc., St. Louis, MO) according to the manufacturer’s protocol. PCR was performed to characterize the native AalNix and the exogenous transgenic AalNix with the primers Nix_i2-F/ Nix_i2-R and the primers Nix_E1F/ strep tag-R, using DreamTaq Hot Start Green PCR Master Mix (2X) (Thermo Fisher Scientific, Waltham, MA, USA) according to the user manual. Males containing the endogenous AalNix and exogenous AalNix genes were mated with wild-type females to established lines.
Wing-length measurement and statistical analysis
Wing-length values were measured for 30 individuals from wild-type females and males and AalNix3&4-♂4-pseudo-males. The wings of one mosquito were removed and placed under a laboratory microscope (connected to a computer and a camera) to take photographs. Image-pro Plus software was used to measure the wing length of adult mosquitoes. The wing length as determined is the distance from the axillary incision to the apical margin (excluding the wing-margin fringe hairs). The average length of a pair of wings from each mosquito was recorded as the wing length.
RT-qPCR analysis of transgenic mosquitos
The relative expression levels of AalNix, Aalbdsx and Aalfru in transgenic mosquitos were quantified by real-time PCR using the SuperReal PreMix Plus kit (SYBR Green) (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer’s instructions. Total RNA was extracted from whole adult transgenic mosquitoes from each phenotype of each line, using TRIzol (Life Technologies, Carlsbad CA, USA) according to the manufacturer’s directions. Each sample contains nine adult mosquitos assessed in triplicate and normalized with AalRpS7 mRNA. The qPCR results were analyzed using the 2−ΔΔCT method.
Mating competition assays
To assess mating competitiveness between wild type and transgenic males (m/m♂; nix/+), five WT males and five transgenic males were placed into a 1.5-L container and allowed to adapt to the environment. A total 10 virgins females previously fed with a blood meal were put into the container to mate. After two day of full mating, the female mosquitoes in each container were placed in a single oviposition cup to lay eggs and these were allowed to hatch. When the hatched larvae reached the third-fourth instar, the were screened with fluorescence microscopy (Nikon SMZ1000) and counted. Six replicate cages were set up and assayed for each line as described.
Statistical analysis
All data were represented as means ± standard deviation (SD) from at least three independent experiments. Analysis of differences between groups were analyzed by unpaired Student’s t-test. One-Way ANOVA of variance was used to evaluate spatiotemporal expression profiles of AalNix. Statistical significance was set at P < 0.05 and was determined using GraphPad Prism Version 8.02 program (GraphPad Software, CA, USA) and IBM SPSS Statistics 20.0 (SPSS Software, IL, USA). Gene expression was normalized using the deltaCT method against AalRpS7 as a reference gene.
Supporting information
(a) AalNix in the isoform1-♂1 strain. (b) AalNix in the isoform2-♂3 strain. (c) AalNix in the isoform4-♂15 strain. Agarose gel electrophoresis of PCR products showing the result of amplifying the flanking regions. Sequence analysis showed that transgenic line insertions are in intergenic regions. The band represented by the asterisk (*) results from incomplete digestion.
(TIF)
(a) The primer pair Nix_i2-F/Nix_i2-R is located in intron 2 and were designed to distinguish the endogenous AalNix from the transgenic Nix. (b) Primer pair Nix_E1F/strep tag-R that span the exon1 and Strep II tag were designed to confirm transgene insertion.
(TIF)
Deformities and masculinization in internal reproductive organs in transgenic female mosquitoes, we define this phenotype of mosquitoes as intersex. External genitalia (top panels), and internal genitalia (bottom panels).
(TIF)
Red font peptide indicates AalNix3&4 intron1. Green front indicates AaeNix Exon2.
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Omar Akbari (University of California San Diego, La Jolla, CA) for providing the plasmid AAEL010097-Cas9 used in this study. AAJ is a Donald Bren Professor.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (81420108024, 31830087), the National Key Research and Development Program of China (2020YFC1200100), and the National Institutes of Health, USA (AI136850) to X-G.C. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lowe S, Browne M, Boudjelas S, De Poorter M. 100 of the world’s worst invasive alien species: a selection from the global invasive species database: Invasive Species Specialist Group Auckland; 2000. [Google Scholar]
- 2.Simmons CP, Farrar JJ, van Vinh Chau N, Wills BJNEJoM. Dengue. The New England Journal of Medicine. 2012;366(15):1423–32. doi: 10.1056/NEJMra1110265 [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Zhou T, Lai Z, Zhang Z, Jia Z, Zhou G, et al. Competence of Aedes aegypti, Ae. albopictus, and Culex quinquefasciatus mosquitoes as Zika virus vectors, China. Emerging infectious diseases. 2017;23(7):1085. doi: 10.3201/eid2307.161528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jentes ES, Poumerol G, Gershman MD, Hill DR, Lemarchand J, Lewis RF, et al. The revised global yellow fever risk map and recommendations for vaccination, 2010: consensus of the Informal WHO Working Group on Geographic Risk for Yellow Fever. The Lancet Infectious Diseases. 2011;11(8):622–32. doi: 10.1016/S1473-3099(11)70147-5 [DOI] [PubMed] [Google Scholar]
- 5.Leparc-Goffart I, Nougairede A, Cassadou S, Prat C, De Lamballerie XJTL. Chikungunya in the Americas. Lancet. 2014;383(9916):514. doi: 10.1016/S0140-6736(14)60185-9 [DOI] [PubMed] [Google Scholar]
- 6.Paupy C, Delatte H, Bagny L, Corbel V, Fontenille D. Aedes albopictus, an arbovirus vector: from the darkness to the light. Microbes and infection. 2009;11(14–15):1177–85. Epub 2009/05/20. doi: 10.1016/j.micinf.2009.05.005 . [DOI] [PubMed] [Google Scholar]
- 7.Hall AB, Basu S, Jiang X, Qi Y, Timoshevskiy VA, Biedler JK, et al. SEX DETERMINATION. A male-determining factor in the mosquito Aedes aegypti. Science. 2015;348(6240):1268–70. Epub 2015/05/23. doi: 10.1126/science.aaa2850 ; PubMed Central PMCID: PMC5026532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu P, Jin B, Li X, Zhao Y, Gu J, Biedler JK, et al. Nix is a male-determining factor in the Asian tiger mosquito Aedes albopictus. Insect Biochem Mol Biol. 2020;118:103311. Epub 2020/01/07. doi: 10.1016/j.ibmb.2019.103311 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Criscione F, Qi YM, Tu ZJ. GUY1 confers complete female lethality and is a strong candidate for a male-determining factor in Anopheles stephensi. Elife. 2016;5:e19281. WOS:000386117500001. doi: 10.7554/eLife.19281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall AB, Papathanos PA, Sharma A, Cheng C, Akbari OS, Assour L, et al. Radical remodeling of the Y chromosome in a recent radiation of malaria mosquitoes. PNAS 2016;113(15):E2114–23. Epub 2016/04/02. doi: 10.1073/pnas.1525164113 ; PubMed Central PMCID: PMC4839409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzywinska E, Dennison NJ, Lycett GJ, Krzywinski J. A maleness gene in the malaria mosquito Anopheles gambiae. Science. 2016;353(6294):67–9. Epub 2016/07/02. doi: 10.1126/science.aaf5605 . [DOI] [PubMed] [Google Scholar]
- 12.Sharma A, Heinze SD, Wu Y, Kohlbrenner T, Morilla I, Brunner C, et al. Male sex in houseflies is determined by Mdmd, a paralog of the generic splice factor gene CWC22. Science. 2017;356(6338):642–5. Epub 2017/05/13. doi: 10.1126/science.aam5498 . [DOI] [PubMed] [Google Scholar]
- 13.Meccariello A, Salvemini M, Primo P, Hall B, Koskinioti P, Dalikova M, et al. Maleness-on-the-Y (MoY) orchestrates male sex determination in major agricultural fruit fly pests. Science. 2019;365(6460):1457–60. Epub 2019/08/31. doi: 10.1126/science.aax1318 . [DOI] [PubMed] [Google Scholar]
- 14.Sawanth SK, Gopinath G, Sambrani N, Arunkumar KPJJob. The autoregulatory loop: A common mechanism of regulation of key sex determining genes in insects. Journal of Biosciences. 2016;41(2):283–94. doi: 10.1007/s12038-016-9609-x [DOI] [PubMed] [Google Scholar]
- 15.Jin B, Zhao Y, Dong Y, Liu P, Sun Y, Li X, et al. Alternative splicing patterns of doublesex reveal a missing link between Nix and doublesex in the sex determination cascade of Aedes albopictus. Insect science. 2021;28(6):1601–20. Epub 2020/11/13. doi: 10.1111/1744-7917.12886 . [DOI] [PubMed] [Google Scholar]
- 16.Hoshijima K, Inoue K, Higuchi I, Sakamoto H, Shimura YJS. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. science. 1991;252(5007):833–6. doi: 10.1126/science.1902987 [DOI] [PubMed] [Google Scholar]
- 17.Ryner LC, Baker BSJG, development. Regulation of doublesex pre-mRNA processing occurs by 3’-splice site activation. Genes & Development. 1991;5(11):2071–85. doi: 10.1101/gad.5.11.2071 [DOI] [PubMed] [Google Scholar]
- 18.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell 1993;74(1):105–14. doi: 10.1016/0092-8674(93)90298-5 [DOI] [PubMed] [Google Scholar]
- 19.Adelman ZN, Tu Z. Control of mosquito-borne infectious diseases: sex and gene drive. Trends in Parasitology. 2016;32(3):219–29. doi: 10.1016/j.pt.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhulst EC, van de Zande L, Beukeboom LWJCoig, development. Insect sex determination: it all evolves around transformer. Current opinion in genetics & development. 2010;20(4):376–83. doi: 10.1016/j.gde.2010.05.001 [DOI] [PubMed] [Google Scholar]
- 21.Lutrat C, Olmo RP, Baldet T, Bouyer J, Marois E. Transgenic expression of Nix converts genetic females into males and allows automated sex sorting in Aedes albopictus. Communications biology. 2022;5(1):210. Epub 2022/03/09. doi: 10.1038/s42003-022-03165-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aryan A, Anderson MA, Biedler JK, Qi Y, Overcash JM, Naumenko AN, et al. Nix alone is sufficient to convert female Aedes aegypti into fertile males and myo-sex is needed for male flight. PNAS. 2020;117(30):17702–9. doi: 10.1073/pnas.2001132117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palatini U, Masri RA, Cosme LV, Koren S, Thibaud-Nissen F, Biedler JK, et al. Improved reference genome of the arboviral vector Aedes albopictus. Genome biology. 2020;21(1):215. Epub 2020/08/28. doi: 10.1186/s13059-020-02141-w ; PubMed Central PMCID: PMC7448346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, et al. Ten things you should know about transposable elements. Genome biology. 2018;19(1):199. Epub 2018/11/21. doi: 10.1186/s13059-018-1577-z ; PubMed Central PMCID: PMC6240941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macias VM, Ohm JR, Rasgon JL. Gene Drive for Mosquito Control: Where Did It Come from and Where Are We Headed? International journal of environmental research and public health. 2017;14(9). Epub 2017/09/05. doi: 10.3390/ijerph14091006 ; PubMed Central PMCID: PMC5615543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh S, Jokhadze G, Duong T, Bostick M, Farmer AAJSBE, Evolution, Design. Optimized In-Fusion cloningsystem demonstrates high efficiency and accuracy of multi-fragment cloning. 2014. [Google Scholar]
- 27.Chen XG, Marinotti O, Whitman L, Jasinskiene N, James AA, Romans P. The Anopheles gambiae vitellogenin gene (VGT2) promoter directs persistent accumulation of a reporter gene product in transgenic Anopheles stephensi following multiple bloodmeals. Am J Trop Med Hyg. 2007;76(6):1118–24. Epub 2007/06/09. 10.4269/ajtmh.2007.76.1118. . [DOI] [PubMed] [Google Scholar]
- 28.Handler AM, Harrell RA 2nd. Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8(4):449–57. Epub 2000/01/15. doi: 10.1046/j.1365-2583.1999.00139.x . [DOI] [PubMed] [Google Scholar]
- 29.Jasinskiene N, Juhn J, James AA. Microinjection of A. aegypti embryos to obtain transgenic mosquitoes. JoVE. 2007(5):e219. doi: 10.3791/219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T, Yang WQ, Xie YG, Liu PW, Xie LH, Lin F, et al. Construction of an efficient genomic editing system with CRISPR/Cas9 in the vector mosquito Aedes albopictus. Insect science. 2019;26(6):1045–54. Epub 2018/10/13. doi: 10.1111/1744-7917.12645 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) AalNix in the isoform1-♂1 strain. (b) AalNix in the isoform2-♂3 strain. (c) AalNix in the isoform4-♂15 strain. Agarose gel electrophoresis of PCR products showing the result of amplifying the flanking regions. Sequence analysis showed that transgenic line insertions are in intergenic regions. The band represented by the asterisk (*) results from incomplete digestion.
(TIF)
(a) The primer pair Nix_i2-F/Nix_i2-R is located in intron 2 and were designed to distinguish the endogenous AalNix from the transgenic Nix. (b) Primer pair Nix_E1F/strep tag-R that span the exon1 and Strep II tag were designed to confirm transgene insertion.
(TIF)
Deformities and masculinization in internal reproductive organs in transgenic female mosquitoes, we define this phenotype of mosquitoes as intersex. External genitalia (top panels), and internal genitalia (bottom panels).
(TIF)
Red font peptide indicates AalNix3&4 intron1. Green front indicates AaeNix Exon2.
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.