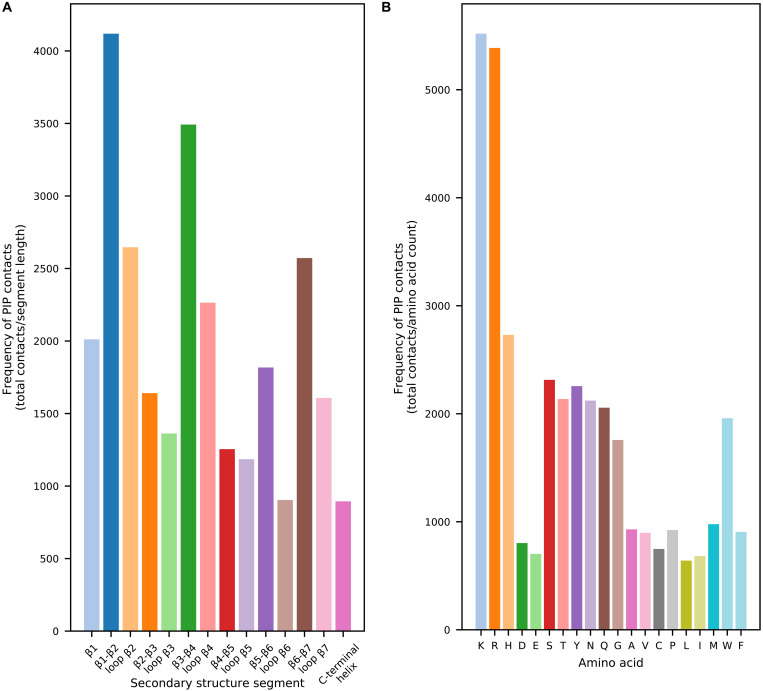
Fig. 4. Secondary structure and amino acid contributions to phosphoinositide contacts.
(A) Frequency with which each of the secondary segments of PH domains made contacts with phosphoinositide headgroups during all simulations. PH domain residues were assigned to 1 of 14 conserved secondary structure units, and contact frequency was calculated by summing contacts for each residue assigned to the secondary structure unit over all PH domains and simulation replicates and dividing by the total number of residues assigned to that secondary structure unit. (B) Frequency with which each amino acid type contacted PIP2 and PIP3 headgroups during all simulations. Frequency was calculated by summing contacts for the amino acid over all simulation replicates of all PH domains and dividing by the total number of occurrences of that amino acid in the simulated sequences.