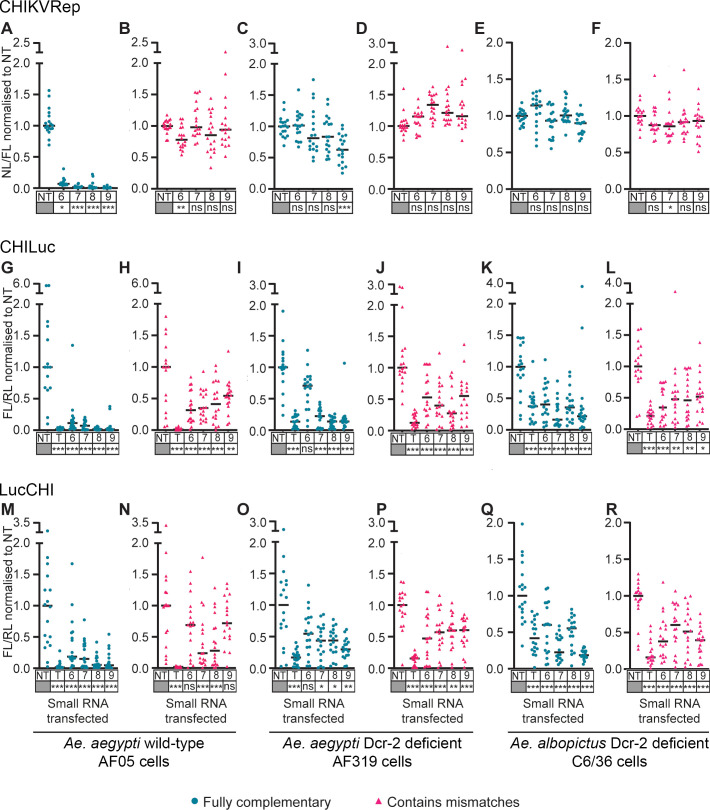
Fig 4.
Effect of selected small RNAs on expression of a CHIKV split replication system and different reporter systems containing viral sequences in Ae. aegypti AF05 (A, B, G, H, M, N) cells, and Dcr-2 deficient Ae. aegypti AF319 (C, D, I, J, O, P) and Ae. albopictus C6/36 (E, F, K, L, Q, R) cells. Plasmids expressing selected shRNA-like small RNAs (Fully complementary, blue) or miRNA-like small RNAs (Contains mismatches, pink) were co-transfected with a split replication system with nanoluciferase (NL) under the control of a CHIKV subgenomic promoter, and a plasmid expressing firefly luciferase (FL) as control for transfection efficiency (A-F), a chimeric CHILuc reporter containing the target sequence within the protein coding region (G-L), or a LucCHI reporter which has the target sequence in the 3’ UTR (M-R). A plasmid expressing Renilla luciferase (RL) as control for transfection efficiency was used in G-R, as well as small RNA targeting FL as positive control (T). Numbers 6 to 9 represent each small RNA tested. Each line represents the median values of NL and FL ratios (NL/FL) or FL and RL ratios (FL/RL) normalized to the non-targeting control (NT). Three independent transfections with at least five replicates each, and each data point represents a replicate. (B, C, E, F, G, H, J, K, N, P-R) Linear mixed effect model, (A, D, I, L, O) Kruskal Wallis, (M) one-way ANOVA. Statistical differences indicate higher silencing efficiency of the siRNA compared to the non-targeting siRNA control. Statistical significance of the silencing effect between each small RNA and the NT is denoted under each small RNA name (*** P<0.001, ** P<0.01, * P<0.05, ns: not significant) (S14–S16 Tables).