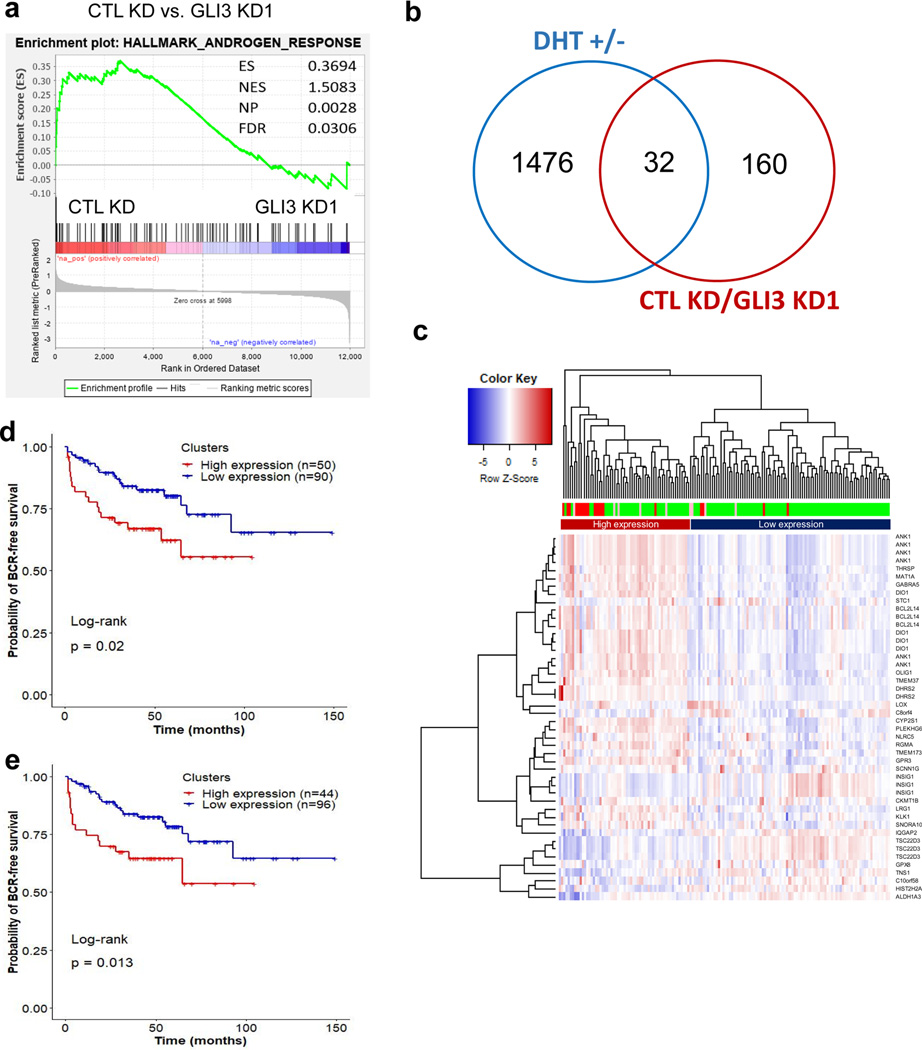
Figure 6. GLI3 and AR co-regulated genes correlate with metastatic CRPC and predict recurrence.
a Gene set enrichment analysis shows significant enrichment of Hallmark Androgen Response genes in control (CTL KD) compared to GLI3 knockdown (GLI3 KD1) LNCaP AI cells. b Venn diagram comparison of genes found significantly upregulated by ≥ 2 fold in androgen replete (+DHT) compared to androgen-depleted (-DHT) LNCaP cells versus genes with significant ≥ 2 fold upregulation in control (CTL KD) compared to GLI3 knockdown (GLI3 KD1) androgen-depleted LNCaP cells. c Heatmap representation of unsupervised hierarchical clustering of 150 tumor samples derived from prostate cancer patients(50) based on log2 (expression) values of 46 transcripts (from 31 androgen and GLI3 co-upregulated genes). One of the 32 genes in Panel b, LGALS8-AS1, was not found in the gene expression data provided by Taylor et al.(50) and hence excluded. The heatmap was plotted using R based ‘gplots’ package with default parameters. Red, green, and pink bars indicate metastatic tumor, primary tumor, and primary tumor samples from patients with observed clinical metastasis, respectively. d Kaplan-Meier plots depicting BCR-free survival of 140 prostate cancer patients(50) stratified by expression clusters from panel (c). e Kaplan-Meier plots depicting BCR-free survival of 140 prostate cancer patients stratified by average expression of 9 transcripts (from 6 genes), which were found significantly (p-value < 0.05) upregulated in 19 metastatic tumors compared to 131 primary tumors. The expression values of the 9 transcripts for each patient were averaged. The patients with expression above and below the overall average expression of all 150 samples were classified as high and low expression groups, respectively.