Figure 6. ESRRA drives the PT differentiation state and protects from kidney disease.
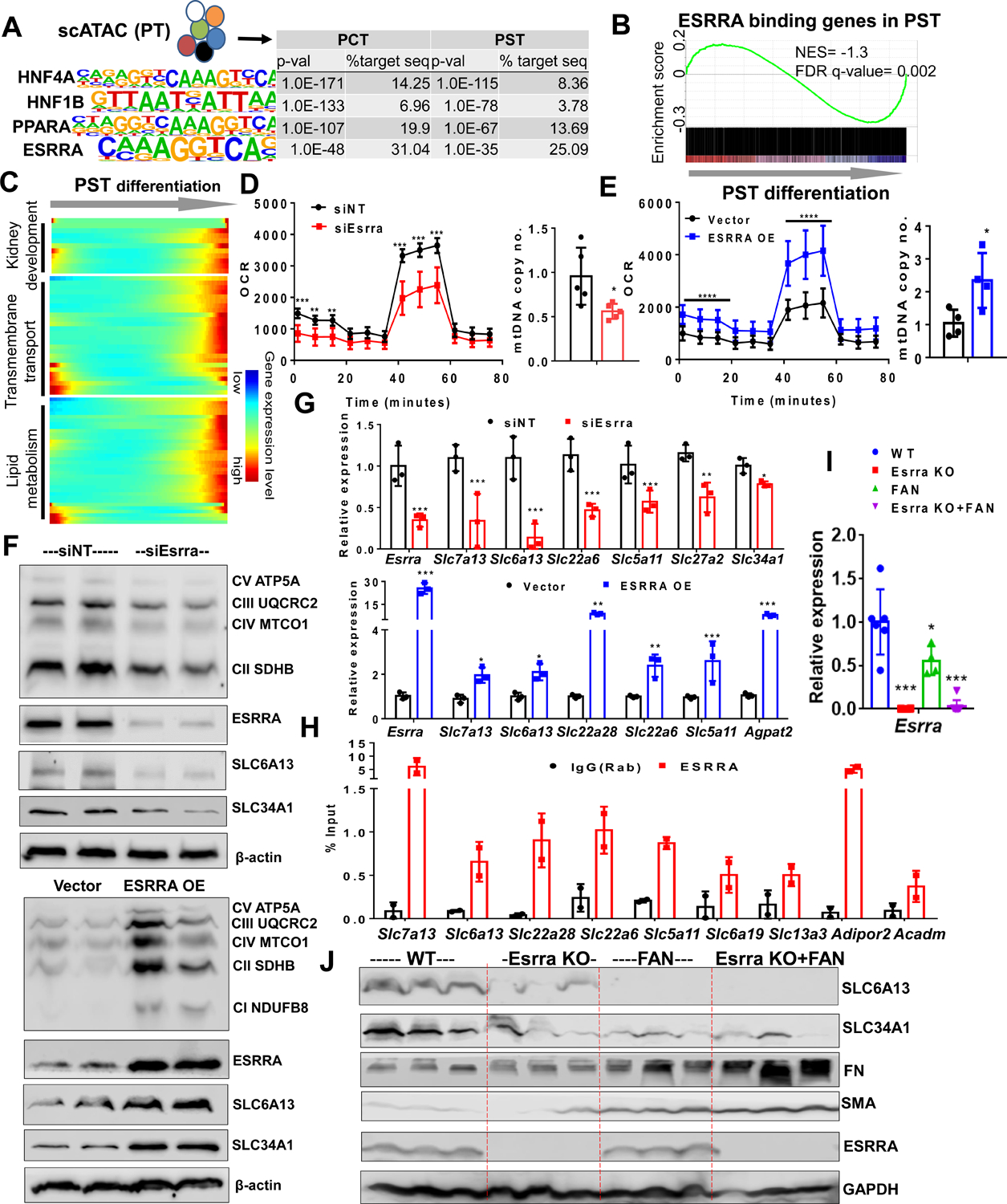
(A) Top transcription factor-binding motifs significantly enriched in PT cell-specific open chromatin regions that are identified from mouse single cell ATAC-sequencing. P-values and percent of target sequences among all open chromatin regions are shown in the table on right.
(B) Gene Set Enrichment Analysis (GSEA) enrichment plot of ESRRA target genes along PST cell differentiation.
(C) Heatmap showing the expression changes of ESRRA target genes along the PST differentiation trajectory (ordered from Figure 4C) grouped by functional annotation (kidney development, transmembrane transport and lipid metabolism).
(D) Oxygen consumption rate (OCR) (pmol/min/μg of protein) and mtDNA copy number (ratio of mtDNA to nuclear DNA) in LTL+ PT cells transfected with non-target siRNA (siNT: black) and ESRRA siRNA (siEsrra: Red) for 2 days. * P < 0.05, ** P < 0.01, *** P < 0.001 vs. siNT.
(E) OCR and mtDNA copy no. in LTL+ PT cells transfected with vector alone (black) and ESRRA expressing vector (ESSRA OE: Blue) for 48 hours. * P < 0.05, ** P < 0.01, *** P < 0.001 vs. vector.
(F) Protein levels of OXPHOS, ESRRA, SLC6A13, and SLC34A1 in LTL+ PT cells transfected with siEsrra (upper panel) or ESRRA OE (lower panel) shown by Western Blot. β-actin was used as loading control.
(G) Relative mRNA levels of Esrra and variety of SLCs markers (Slc7a13, Slc6a13, Slc22a6, Slc5a11, Slc27a2, and Slc34a1) in LTL+ PT cells transfected with siNT, siEsrra (red) and ESRRA OE plasmid (blue). * P < 0.05, ** P < 0.01, *** P < 0.001 vs. siNT/vector.
(H) ChIP-qPCR of ESRRA showed enrichment in genes including SLCs markers (Slc7a13, Slc6a13, Slc22a28, Slc5a11, Slc6a19, and Slc13a3) and metabolic genes (Adipor2 and Acadm) in LTL+ PT cells compared to IgG control.
(I) Relative gene expression of Esrra measured by qRT-PCR in kidneys of wild type, Esrra knock-out mice, sham or FAN treated mice. * P < 0.05, ** P < 0.01, *** P < 0.001 vs. WT.
(J) Protein levels of SLC6A13, SLC34A1, FN, SMA, and ESRRA in kidneys of wild type, Esrra knock-out mice, sham or FAN treated mice were analyzed by Western Blot. GAPDH was used as loading control.
